Hepatocellular carcinoma (HCC) is the third most frequent cancer of digestive tract tumors in Peru, with a high mortality rate of 17.7 per 100,000 inhabitants. A significant number of HCC cases in Peru do not follow the classic clinical epidemiology of the disease described in other parts of the world. Those patients present with a distinct transcriptome profile and a singular tumor process, suggesting a particular type of hepatocarcinogenesis in a portion of the Peruvian population. Our aim was to understand the clinical and biologic involvement of the epigenetic profile (methylation) and gene expression (transcriptome) of HCC in Peruvian patients.
MethodsHCC and liver transcriptome and DNA methylation profiles were evaluated in 74 Peruvian patients.
ResultsWhen grouped by age, there was greater DNA methylation in younger patients with HCC but no differences with respect to the transcriptomic profile. A high prevalence of the hepatitis B virus (HBV) (>90%) was also observed in the younger patients with HCC. Enrichment analyses in both molecular profiles pinpointed PRC2 as an important molecular effector of that liver tumor process in Peruvian patients.
ConclusionHCC in Peruvian patients has a unique molecular profile, associated with the presence of HBV, as well as overall DNA hypermethylation related to undifferentiated liver cells or cellular reprogramming.
En Perú, el carcinoma hepatocelular (CHC) ocupa el tercer lugar en incidencia entre los tumores del sistema digestivo, y tiene una alta tasa de mortalidad, 17.7 por 100 000 habitantes. La mayoría de los casos reportados no presentan la epidemiología clínica clásica del CHC observado en otras partes del mundo. Además, se ha identificado que estos pacientes presentan un perfil transcriptómico distinto y un proceso tumoral singular, sugiriendo un proceso particular de hepatocarcinogénesis en una fracción de la población peruana.
ObjetivoEl presente estudio busca comprender la implicancia clínica y biológica del perfil epigenético (metilación) y de expresión de los genes (transcriptómico) del CHC en los pacientes peruanos.
MétodosSe evaluó el perfil de transcriptómico y de metilación de ADN de hígado y CHC en 74 pacientes peruanos.
ResultadosEl agrupamiento por edades mostró una mayor metilación del ADN en los pacientes jóvenes con CHC, en contraste no se observaron diferencias en el perfil transcriptómico. Adicionalmente, también se evidenció una alta prevalencia del virus de la hepatitis B (VHB) (>90%) en los pacientes jóvenes con CHC. El análisis de enriquecimiento en ambos perfiles moleculares demostró que PRC2 es posiblemente uno de los principales actores moleculares en este proceso tumoral hepático en pacientes peruanos.
ConclusiónEl CHC peruano presenta un perfil molecular único, asociado a la presencia del VHB, y con una hipermetilación global del ADN asociado a células hepáticas indiferenciadas o a una reprogramación celular.
Liver cancer is currently considered a worldwide public health problem, due to its high mortality rate, poor prognosis, and frequent recurrence.1 It is the sixth type of cancer with the highest incidence, and the fourth in mortality, with more than 800,000 cases and 780,000 deaths across the globe. Between 75 and 85% of the cases of primary liver cancer are hepatocellular carcinoma (HCC).2
HCC is considered an aggressive cancer. Its main risk factor is cirrhosis of the liver (80–90%), caused by high levels of alcohol consumption, chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infections, and other metabolic conditions, such as nonalcoholic steatohepatitis.3,4 There is also a risk from environmental factors, such as exposure to mycotoxins (aflatoxin B1),5 pesticides,6 and metalloids (arsenic).7 The multiple factors that give rise to HCC involve a wide variety of molecular patterns associated with the DNA level and gene transcription. In 2015, Zucman-Rossi et al. proposed the first molecular classification consensus, with two main groups of HCC: i) proliferative and ii) nonproliferative.8 Those groups are characterized by different mechanisms of liver carcinogenesis (e.g., risk factors, gene expression signatures, and specific activation pathways). It should also be mentioned that, at the epigenetic level, there is generally an overall loss of DNA methylation (hypomethylation) in HCC, similar to that observed in other carcinomas.9,10
In South America, the high incidence of HCC observed in Peru places it in second place for that type of cancer, surpassed only by Bolivia.11 Nevertheless, a very significant number of Peruvian patients with HCC have not had a previous development of cirrhosis.12–15 In the last 10 years, the Peruvian population with HCC has been shown to present particular characteristics at different levels (epidemiologic, clinical, pathologic, genetic, transcriptomic, epigenetic, etc.), that have not been previously reported at such a magnitude in other parts of the world. The most relevant clinical characteristics of the patients with HCC seen at the Instituto Nacional de Enfermedades Neoplásicas (INEN) are: i) a bimodality regarding age, with a median age of 44 years (two age groups: ≤44 and >44), ii) extremely large tumor presentation (>10 cm) in most of the patients, iii) no presentation of symptoms associated with cirrhosis in approximately 90% of the patients, and iv) extremely high levels of alpha-fetoprotein (AFP).12,16
From the perspective of genetics and molecular biology, the DNA mutation profile in Peruvian HCC consists mainly of insertions and deletions (indels), whereas the mutation spectrums of HCC in patients from other parts of the world, as described in the literature, are largely transitional and transversional.13 The transcriptomic profile of Peruvian HCC is characterized by a mixed molecular identity, with both proliferative and nonproliferative traits.15 Lastly, epigenetic studies of HCC in Peruvian patients have shown an overall increase in DNA methylation (hypermethylation), once again, in contrast with that reported in other regions (hypomethylation).9,10,15
Nevertheless, certain clinical genetic and molecular characteristics of HCC in the Peruvian population have not been studied. Therefore, the present study examines the demographic, clinical, and biologic aspect of the transcriptomic and epigenetic profiles of Peruvian HCC, utilizing a different analytic focus from that previously reported.
Material and methodsStudy design and populationThe transcriptomic (n = 39) and methylation (n = 70) profile data, as well as the clinical and pathologic characteristics of Peruvian patients diagnosed with HCC and programmed for liver resection, within the time frame of 2006 and 2016 at the INEN, were obtained from a previous study and downloaded from the Gene Expression Omnibus (GEO) database.15 In brief, none of the samples collected came from patients that had undergone chemotherapy before liver resection, in accordance with the recommendations of the European Association for the Study of the Liver (EASL).17 The samples were collected in pairs; an HCC tissue sample and an adjacent nontumor liver tissue (NTL) sample extracted from the surgical margin (≥2 cm). Approximately 50 mg of each tissue type (HCC and NTL) were flash frozen and stored at −150 °C in the Tumor Tissue Bank of the INEN before analysis. The variables considered in the present study included demographic, laboratory, histologic, and HBV infection data. Table S1 shows the details of each variable.
Preoperative survival prediction nomogramThe patients were classified according to the survival prognosis nomogram for tumors >10 cm.18 The preoperative variables of that nomogram are: i) presence of cirrhosis, ii) multiple tumor lesions, iii) macroscopic vascular invasion, and iv) spontaneous tumor rupture. From the final score, the samples were classified into three survival groups: G1, with a high probability of long-term survival (0 points); G2, with a 60% probability of recurrence within 12 months (between one and 100 points); and G3, with the highest postoperative mortality (>100 points).
Detection of hepatitis B virus DNAHBV DNA detection in tissues was carried out as previously described.19 In short, genomic DNA was extracted using the phenol-chloroform method, and quantification was done, using the Qubit® (Invitrogen) dsDNA kit. All samples were examined for detecting the presence of HBV DNA, utilizing the PCR QX200™ Droplet Digital™ (Bio-Rad) system. HBV sequencing was performed using the BigDye™ Terminator v3.1 (Applied Biosystems) sequencing kit and HBV phylogeny was determined using the two-parameter Kimura model and the neighbor joining method in version 7 of the Molecular Evolutionary Genetics Analysis software (MEGA, University Park, PENN, USA) The sequencing data can be downloaded from the European Bioinformatics Institute (EBI) site, with the access code: PRJEB21100 (https://www.ebi.ac.uk/ena/data/view/PRJEB21100).
Transcriptome analysisThe transcriptome profile was analyzed from a group of Peruvian HCC samples obtained from the Gene Expression Omnibus database (GEO, Bethesda, MD, USA), with codes GSE136247 and GSE111580.15 The transcriptome of those samples was obtained using GeneChip™ Human Transcriptome Array 2.0 microarrays and the GeneChip™ WT PLUS Reagent Kit (Applied Biosystems, Waltham, MA, USA). The transcriptomic data were normalized through the Robust Multichip Average (RMA) and the lot effect was corrected using the ComBat algorithm of the limma package in R (R language).20 The matrix (genes x samples) was then collapsed using the collapse_genes-09 (Fred’s Softwares, Toulouse, France) algorithm. The differentially expressed genes (DEGs) were obtained, using the R limma package, with a corrected p value (q value) of 0.05, following the recommended workflow.20 In addition, the 5000 most variable genes in the HCC tissue were identified, using standard deviation. Heat maps based on unsupervised hierarchical clustering were created, using the pheatmap R package. Identification of the metabolic pathways of interest was carried out using the Sample Enrichment Score (SES) algorithm and the KEGG, Reactome (reactome_2018803), Gene Ontology-Cellular Component (C5_CC), and MSigDB Hallmarks (H) databases, compiled by Fred’s Softwares.21 The source codes and databases of Fred’s Software are available at: https://sites.google.com/site/fredsoftwares/products.
Methylome analysisThe methylation profile of the Peruvian HCC samples, with codes GSE136319 and GSE136380, were obtained from the GEO database. The methylome of the samples was obtained, using the Infinium HumanMethylation450 K and MethylationEPIC BeadChip (Illumina, San Diego, CA, USA) microarrays. The differentially methylated positions (DMPs) were identified, using the minfi R package, with a q value of 0.05.15 The criteria considered for the methylation analysis were: mean Δβ between HCCs and NTL >5% (for the hypermethylated positions) or ≤ 5% (for the hypomethylated positions). The 5000 positions with the most variable methylation were identified through calculating the standard deviation and the classification, regarding the association with “CpG islands” and “gene structure”, was carried out according to the Illumina definition. The heat maps based on unsupervised hierarchical clustering were created using the pheatmap R package. The SES algorithm was utilized to identify the metabolic pathways of interest and the genes associated with the CpGs of interest, according to the Illumina definition, were used.
Statistical analysisThe multiple factor analyses (MFAs) and principal component analyses (PCAs) were carried out in R, using the FactoMineR package.22 Variables with incomplete data were handled with the imputeMFA function of the missMDA R package. In brief, as recommended in the FactoMineR user guide, an MFA was applied, given that our data had numerical and categorical variables. The MFA was done, according to the recommendations established in the package. All the groups were considered relevant variables and no supplementary data were incorporated in the analysis. In addition, the t-test and chi-square test were done in R. Statistical significance was unilaterally or bilaterally evaluated, depending on the circumstances. All the figures presented herein were created in R and GraphPad Prism v8 (GraphPad Software, Boston, MA, USA).
Ethical considerationsAll the samples included in in the present study came from volunteer patients that accepted the use of their samples and data (in anonymity) for research. The project (code INEN 10-05) was reviewed and approved by the Institutional Committee on Ethics in Research of the INEN.
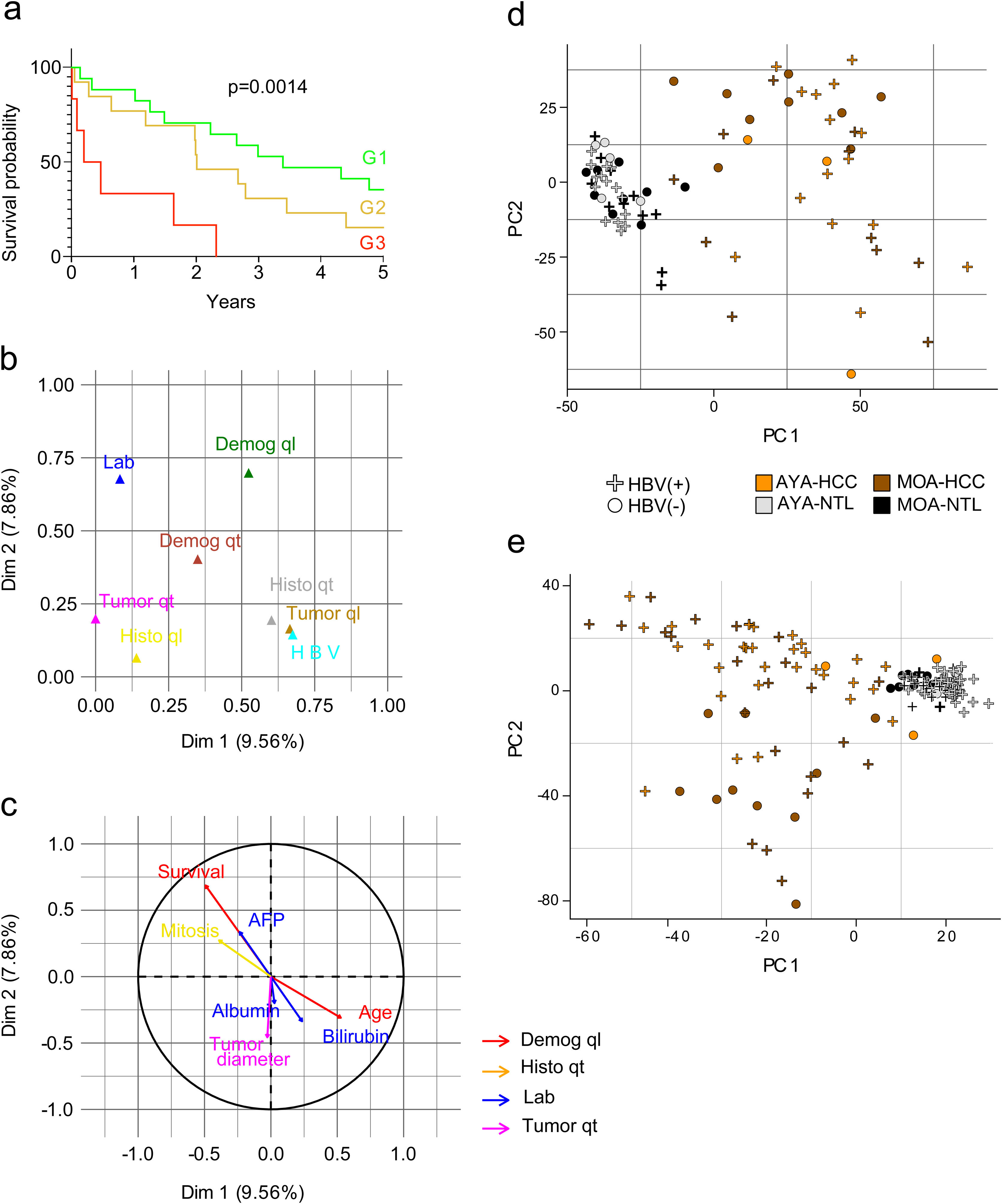
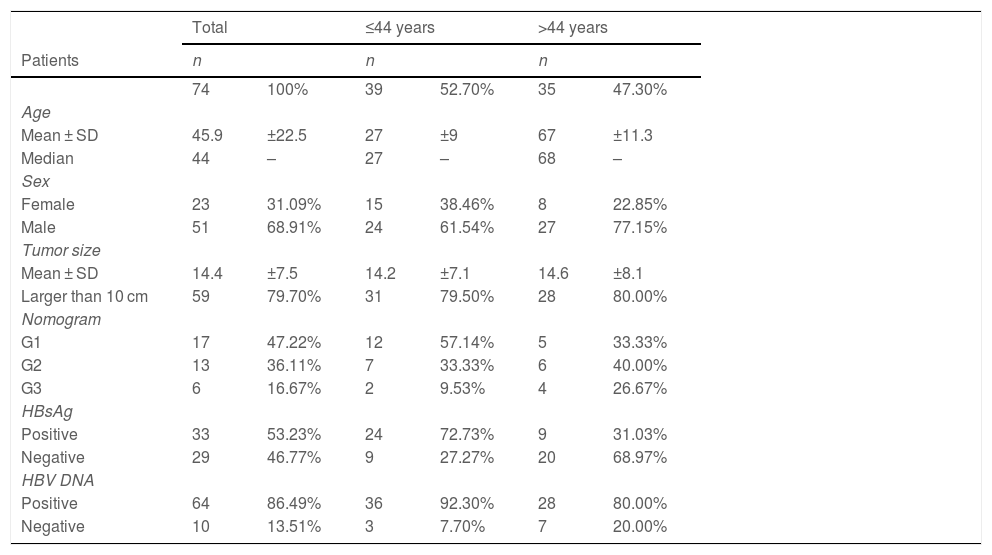
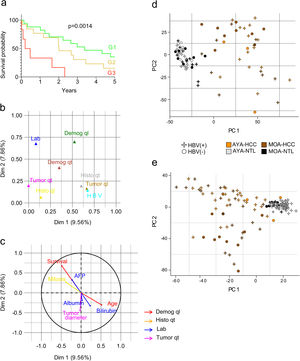
ResultsAge-related clinical and demographic variables of the Peruvian patients with HCCSeventy-four Peruvian patients with HCC that underwent liver resection at the INEN were included in the study. Mean patient age was 45.9 years. In accordance with the median age (44 years), the patients ≤44 years of age were classified as adolescents and young adults (AYAs) and patients >44 years of age were classified as middle or older age individuals (MOAs). The AYA group accounted for more than 50% (n = 39) of the study population with HCC, with a mean age of 27 years, and the MOA group (47.3%; n = 35) had a mean age of 67 years. The male-to-female ratio for the entire patient cohort was two males for each female (2:1) but was double (4:1) in the MOA group. Mean tumor diameter was 14.4 cm (AYAs 14.2 cm and MOAs 14.6 cm; p > 0.05) and the large majority of patients (∼80%) presented with a tumor > 10 cm in diameter. The survival prognosis nomogram for tumors >10 cm,18 was retrospectively applied to 36 patients (21 AYAs and 15 MOAs), and 83% of those patients were in the G1 (n = 17) and G2 (n = 13) groups, both of which had a higher survival probability, compared with the G3 (n = 6) group, in which all the patients died before reaching the three-year postoperative period (Fig. 1a). The Hepatitis B surface antigen (HBsAg) was detected in 53% of the patients (72% of AYAs and 31% of MOAs), whereas HBV DNA was detected in 86% of the study population with HCC; the only sub-genotype identified was F1b. Lastly, the AYA and MOA groups accounted for 92 and 80% (p > 0.05) of the cases with viral DNA, respectively (Table 1).
Evaluation of the clinical and demographic variables in Peruvian patients with HCC. (a) G1 (n = 17), G2 (n = 13), and G3 (n = 6) survival curves of 36 patients, classified according to nomogram. As a result of the MFA, the main components are shown in the graph: (b) the contribution of each group of variables, and (c) the most relevant variables within each group, for the 2 most relevant dimensions (Dim. 1 and 2). Lab (blue); Demog (green and red); HBV (light blue); Histo (yellow and grey); Tumor (brown and fuchsia). Qualitative (ql) and quantitative (qt) data. The PCA of (d) gene expression and (e) DNA methylation data of Peruvian patients, according to HBV infection, age (AYAs and MOAs), and tissue type (HCC and NTL).
Clinical-pathologic description of the HCC study population.
| Total | ≤44 years | >44 years | ||||
|---|---|---|---|---|---|---|
| Patients | n | n | n | |||
| 74 | 100% | 39 | 52.70% | 35 | 47.30% | |
| Age | ||||||
| Mean ± SD | 45.9 | ±22.5 | 27 | ±9 | 67 | ±11.3 |
| Median | 44 | – | 27 | – | 68 | – |
| Sex | ||||||
| Female | 23 | 31.09% | 15 | 38.46% | 8 | 22.85% |
| Male | 51 | 68.91% | 24 | 61.54% | 27 | 77.15% |
| Tumor size | ||||||
| Mean ± SD | 14.4 | ±7.5 | 14.2 | ±7.1 | 14.6 | ±8.1 |
| Larger than 10 cm | 59 | 79.70% | 31 | 79.50% | 28 | 80.00% |
| Nomogram | ||||||
| G1 | 17 | 47.22% | 12 | 57.14% | 5 | 33.33% |
| G2 | 13 | 36.11% | 7 | 33.33% | 6 | 40.00% |
| G3 | 6 | 16.67% | 2 | 9.53% | 4 | 26.67% |
| HBsAg | ||||||
| Positive | 33 | 53.23% | 24 | 72.73% | 9 | 31.03% |
| Negative | 29 | 46.77% | 9 | 27.27% | 20 | 68.97% |
| HBV DNA | ||||||
| Positive | 64 | 86.49% | 36 | 92.30% | 28 | 80.00% |
| Negative | 10 | 13.51% | 3 | 7.70% | 7 | 20.00% |
HBsAg: hepatitis B surface antigen; HBV: hepatitis B virus; SD: standard deviation.
For the MFA, the variables were organized into eight groups: clinical laboratory data (Lab), qualitative demographics (Demog ql), quantitative demographics (Demog qt), cases associated with HBV infection, qualitative histologic (Histo ql) variables, quantitative histologic (Histo qt) variables, qualitative tumor-associated (Tumor ql) variables, and quantitative tumor-associated (Tumor qt) variables (supplementary Table S1). The MFA, utilizing all the clinical and demographic data of the patients with HCC, showed that the groups of the Demog ql, Histo ql, Histo qt, Lab, and Tumor qt variables were the greater contributing parameters (Fig. 1b). The variables within those groups with higher relevance were age, survival, number of mitotic cells (mitosis), AFP, tumor diameter, total bilirubin, and albumin, according to the MFA (Fig. 1c).
Due to the high HBV infection rate, a PCA was done to determine the effect of HBV on gene expression and methylation. In the two settings, there was a higher number of AYA patients infected with HBV, showing that HBV acts as an age-related confounding factor (Fig. 1d, e).
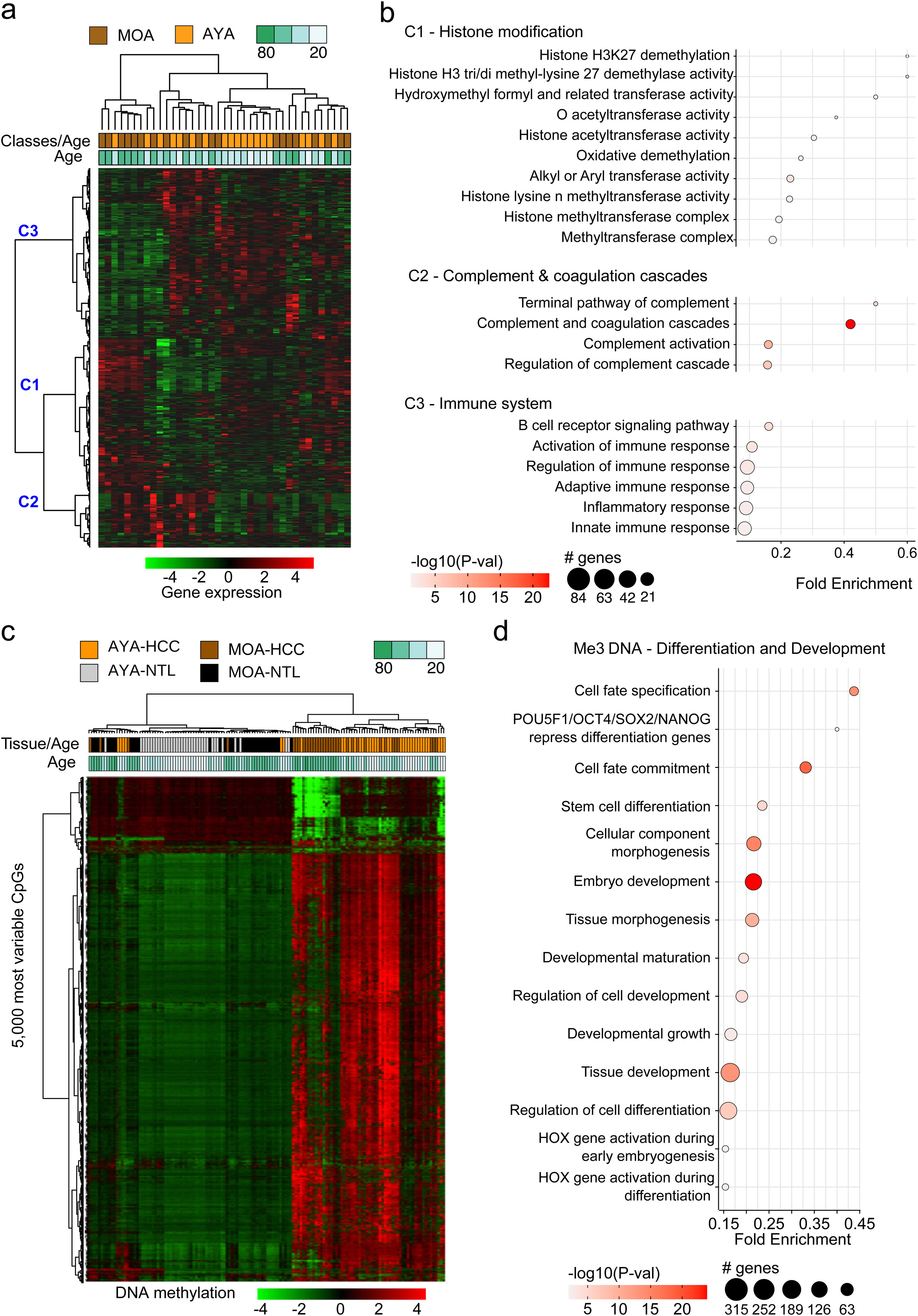
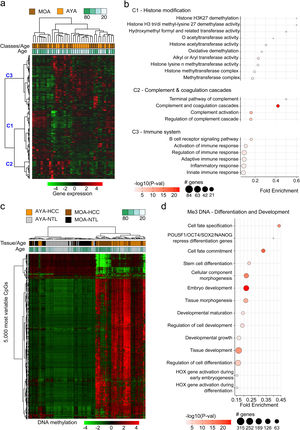
The transcriptomic profile of HCC in Peru was not age-relatedThe transcriptomic profiles in HCC and NTL from a total of 39 patients (19 AYAs and 20 MOAs) were obtained from previously published data.15 The evaluation of the 5000 most variable genes in HCC showed no significant difference between the two age groups (p > 0.05) (Fig. 2a). However, an unsupervised hierarchical cluster analysis of the 5000 most variable genes in the HCC tissue showed the genes were divided into three large groups (C1, C2, and C3). The genes in C1 were mainly associated with expression regulation through histone modification, whereas the majority of genes in C2 participated in the complement cascade and coagulation cascade, and the genes in C3 participated in the immune response. Other less representative pathways were hormone regulation (C1), retinol and lipid metabolism (C1), and DNA repair (C2) (Fig. 2b).
Analysis of the transcriptomic and DNA methylation profiles of Peruvian patients with HCC. 5000 genes (a,b) and 5000 most variable CpGs (c,d) were identified. Utilizing (a) the expression (AYA = 19, MOA = 20) and (c) DNA methylation levels (AYA = 39, MOA = 35) heat maps were created based on an unsupervised hierarchical cluster. In (a), three large gene groups (C1, C2, and C3) stand out. The pathways with greater significance and fold enrichment (b) of the 5000 genes, and (d) of the 5000 most variable CpGs are represented in a dispersion graph. Fold enrichment is the number of genes from the list of interest that are found on the pathway evaluated.
The methylation profiles of HCC and NTL from 70 patients (36 AYAs and 34 MOAs) were obtained from a previous study.15 Similar to the transcriptome analysis, the 5000 most variable CpGs were identified and unsupervised hierarchical clustering was performed. In contrast to the NTL, there was an overall increase in DNA methylation (hypermethylation) in the Peruvian HCC, with greater intensity in the younger patients (Fig. 2c). Those CpGs were also mainly associated with the genes involved in cell differentiation, with methylation marks of lysine residue at N-terminal position 27 of the H3 histone protein (H3K27 [me3]) and targets of the Polycomb Repressive Complex 2 (PRC2) (Fig. 2d).
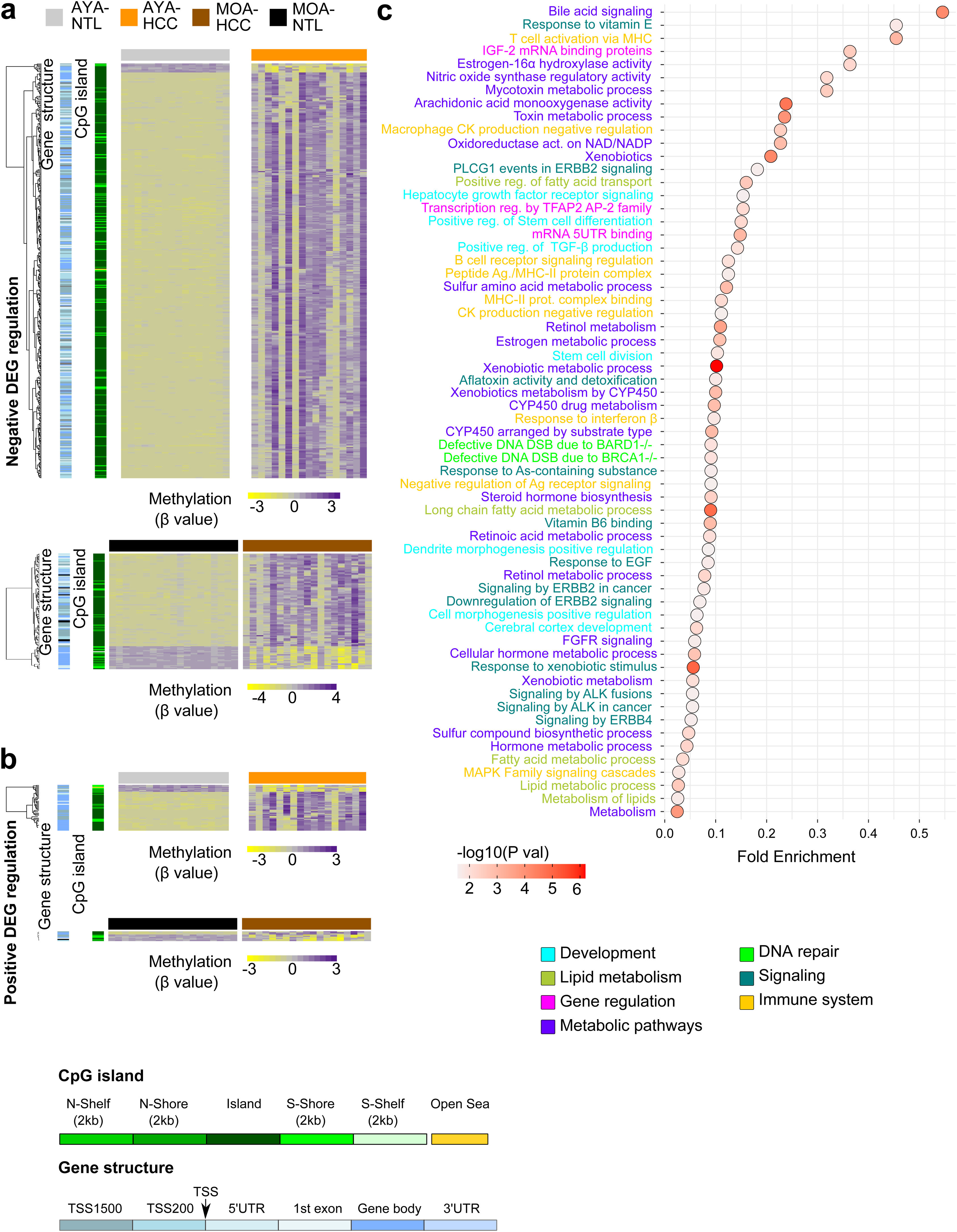
The transcriptome and methylation profile integration underlines cell differentiation and immune system processes in Peruvian HCC patientsTo better understand CpG DNA methylation, a heat map was created on the positively and negatively regulated genes.15 The regions of gene structure and CpG islands were considered independently. The CpGs associated with negatively regulated genes (the majority hypermethylated) were mainly detected on the CpG islands, within the TSS 200 and TSS 1500 promotor regions, whereas the CpGs associated with positively regulated genes (the majority hypomethylated) were located at regions unrelated to the CpG islands in the gene body (Fig. 3a, b). Genomic integration in HCC also revealed a predominance of the pathways related to cell metabolism, but pathways related to cell differentiation and the immune system stood out, as well.
Integrated analysis of the transcriptome and DNA methylation profiles of Peruvian patients with HCC. Heat maps based on an unsupervised hierarchal cluster to show the DNA methylation levels (β value) of (a) the negatively and (b) positively regulated differentially expressed genes (DEGs) in the AYA (n = 39, upper panel) and MOA (n = 35, lower panel) groups. (a,b) The left dendrogram shows the annotation of the CpG island and gene structure. (c) The fold enrichment pathways of greater significance associated with the integrated analysis of gene expression and DNA methylation are illustrated, utilizing a dispersion graph. The fold enrichment is the number of genes on the list of interest that are found on the pathway evaluated.
Lastly, the differences between the groups defined according to the survival prognosis nomogram for tumors >10 cm, at the transcriptomic and DNA methylation levels, were explored. Only 36 patients (17 for the transcriptomic profile and 36 for the DNA methylation profile) had the preoperative variables necessary for being evaluated. A total of 17 patients obtained 0 points, and so were classified as G1, whereas 13 patients obtained between 1–100 points and 6 obtained >100 points, and so were classified as G2 and G3, respectively (Table 1). According to that classification, 47% of the patients had a high probability of long-term survival, greater in those ≤44 years of age (12 AYAs and 5 MOAs), 36% had a high probability of recurrence within the first 12 months (7 AYAs and 6 MOAs), and 17% had a high probability of postoperative death (2 AYAs and 4 MOAs). Despite those clinical differences, the analyses showed no significant differences at the transcriptomic or DNA methylation levels, between the three groups; thus, those analyses were not studied in more detail. Nevertheless, more than 50% of the patients ≤ 44 years of age (57%) had a high probability of long-term survival.
DiscussionIn the present study, we describe the relation between the clinical and pathologic characteristics of Peruvian patients with HCC and a divergent molecular profile that has not been described elsewhere. The presence of occult HBV infection increases the frequency of HBV in cases of HCC in the Peruvian population and is the main risk factor for developing HCC. Even though the bimodality associated with age could suggest particular characteristics for each age group (AYAs and MOAs), such characteristics were not shown in the transcriptomic profile. However, overall DNA hypermethylation in the Peruvian population with HCC was more intense in the AYAs. That strong pattern of hypermethylation in Peruvian patients with HCC could be the characteristic profile of HCC in Peru.
Previous studies by our research group have systematically characterized HCC in the Peruvian population in different areas of study. The combination of those findings has shown that there are characteristics in a significant number of Peruvian patients that are unique to that population.12,18,19,23,24
As expected, a balanced bimodality was observed between AYAs and MOAs, coinciding with previous HCC cohorts analyzed in Peru,12,13 and distribution by sex was balanced, compared with other regions.2 HCC in AYAs is generally the fibrolamellar subtype and is considered a rare type of presentation in some regions, especially in Asia.25 Cases of HCC in AYAs are much more common in the Americas and are mainly associated with HBV infection, without presenting the characteristics of the fibrolamellar subtype,14,26,27 and the percentage reported in Pero is much higher than in other regions of the Americas. The presence of viral DNA (HBV) has been found in more than 85% of the total number of cases of HCC, whereas it was 90% in AYAs.24,28
Even though it is known that high viral loads of chronic HVB infection is one of the main causes of hepatocarcinonogenesis,29 studies in the Peruvian population with HCC have shown a low viral load that could function in synergy, in the presence of other tumorigenic factors.19 In addition, the present study has reported a large quantity (>40%) of cases of occult HBV infection in HCC tumor tissue and NTL, detected only through ultrasensitive molecular methods (such as digital droplet PCR).19,26 The majority of the samples with occult HBV were negative for HBsAg, underlining the need for implementing more sensitive methods for HBV detection in Peru. Because HBV infection is a factor associated with HCC, its role in the development of the disease would be better understood by knowing the true prevalence of HBV in the Peruvian population.
Another particularity of HCC in Peruvian patients is the extremely large tumor size at diagnosis. Approximately 80% of patients present with a tumor larger than 10 cm in diameter, as has been previously reported.13,14,16 Due to the lack of clinical algorithms that aid in predicting the survival of those patients following liver resection, Ruiz et al., in 2021, described a nomogram capable of determining survival in patients with massive tumors.18 Utilizing that algorithm, the survival group of 36 patients was determined (Table 1) and a comparison of their gene expression and DNA methylation profiles was carried out, to find a molecular signature that was characteristic of each group. Unfortunately, no significant differences were found between the three survival groups, showing that HCC in the Peruvian population is a unique entity, or one with little molecular variability.15
In the present study, there were no significant differences in the transcriptomic profile between the AYAs and MOAs, even when another previously described method of analysis was applied.15 Enrichment analyses enabled the identification of groups of pathways related to expression regulation, through lysine 27 modification on histone 3 (H3K27). H3K27 modification is catalyzed by the multisubunit histone-modifying enzyme complex, known as PRC2. Our results concur with those described by Cerapio et al., who showed that PRC2 might be one of the main molecular actors involved in HCC carcinogenic cell reprogramming in Peruvian patients.15 The molecular mechanisms of carcinogenesis in that new HCC subtype have not been described in the literature.
The evaluation of the methylation profile confirms the DNA hypermethylation phenomenon, previously reported in Peruvian patients.15 Those results differ from findings in other regions, in which HCC presents an overall hypomethylation profile, albeit the literature describes focal hypermethylation islands in driver genes.9,10 The overall hypermethylation profile seen in Peruvian HCC has only been reported in induced pluripotent stem cells (iPSCs) that have the capacity to reprogram cells.30 That biologic phenotype concurs with the finding that the 5000 most variable CpGs are associated with genes involved in cell differentiation and development, as described by Cerapio et al. and observed in the present study.15
The integration of the transcriptomic and DNA methylation profiles in Peruvian HCC showed a reduction in gene expression, most likely due to DNA hypermethylation, a profile similar to that of embryonic cells.15,30 Nevertheless, to corroborate that hypothesis, comparative studies must be conducted on cells in differentiation processes to compare them with the molecular profile of HCC in the Peruvian population.
In conclusion, the present study describes HCC with unique characteristics not previously reported in other regions. HBV infections are highly prevalent but can remain undetected by routine diagnostic methods, signifying the need for implementing more sensitive methods. The integration of transcriptomic and DNA methylation profile data paves the way for future detailed studies of the HCC methylation profile, with characteristics of undifferentiated cells or those in the process of differentiation.
Conflict of interestThe authors declare that there is no conflict of interest.
Financial disclosureThe authors declare that this work was supported by the ITMO Cancer of the French National Alliance for Sciences of Life and Health (Aviesan) and the French National Cancer Institute (INCa) with funds administered by the French National Institute of Health and Medical Research (Inserm) (grant 21CD025-00) and the program of International Mixed Laboratories (LMI) of the French Institution for Research Development (IRD). J.C.M. declares financial support of the Proyecto Concytec – World Bank“Mejoramiento y Ampliación de los Servicios del Sistema Nacional de Ciencia Tecnología e Innovación Tecnológica” 8682-PE, through its executive unit ProCiencia (08-2018-FONDECYT/BM-Doctoral Programs in General and Strategic Areas). J.P.C. was the beneficiary of a postdoctoral grant from the Agencia Nacionale de Investigación de Francia (ANR) within the framework of the Laboratories of Excellence Program 11-LABX-0068.