Introduction
Infection by Hepatitis C Virus (HCV) causes chronic liver disease and is a worldwide health problem, affecting 3% of the world's population.1,2 It has been reported that cytokines play key roles in the response to viral infections and a number of them have been associated with the pathogenesis of HCV infection.3 Tumor necrosis factor alpha (TNF-α) is a pro-inflammatory cytokine that has been associated with HCV infection.4 Several studies have shown that serum and messenger ribonucleic acid (mRNA), levels of TNF-α and soluble TNF receptors (sTNFR) may correlate with histological severity and/or lack of response to therapy in chronically ill HCV-infected patients.5-7 However, these data have been inconsistent because Falasca et al.8 reported that TNF-α and interleukin-2 (IL-2) serum levels were higher in hepatitis B virus patients in comparison to HCV positive patients (P <0.001). Additionally, the level of sTNFR has been proposed as a good indicator of TNF production; therefore the sTNFR level is more valuable for monitoring the degree of TNF-α system activity than the TNF-α level.9
G to A polymorphic sequence at position -308 (TNF1 to TNF2 allele) and -238 (TNFG to TNFA allele) in the TNF-α promoter has been shown to influence TNF-α gene expression.10,11 Moreover, these polymorphisms have been associated with clinical characteristics of chronic hepatitis C.12,13 However, conflicting data with regard to the association between TNF-α promoter polymorphisms and the progression of chronic hepatitis C or response to interferon (INF) therapy have been reported.14,15 Therefore, the aim of this study was to analyze the prevalence of the -308 and -238 TNF- polymorphisms in a group of Mexican HCV-infected patients and control subjects not related to the patients.
Material and methods
Patients and Control Subjects
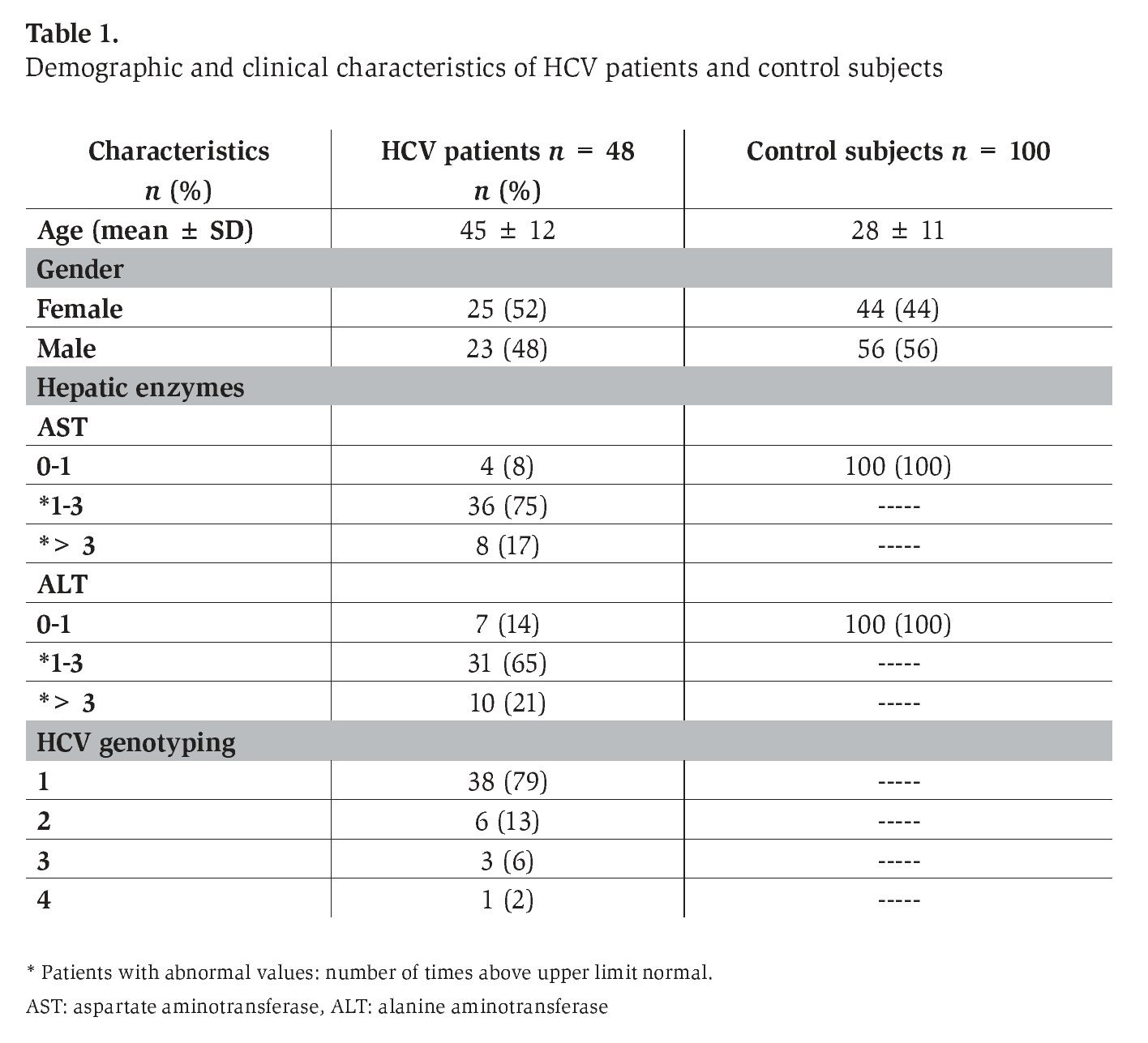
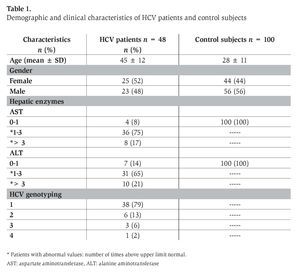
We studied 48 HCV-infected patients (25 women and 23 men, mean age 45 ± 12 years) being treated at the Infectious Diseases Service and Liver Unit at the University Hospital in Monterrey and community-based out-reach facilities. HIV/HCV co-infected patients and pregnant woman were not included in this study. The aforementioned public hospital is the largest in northeastern Mexico with patients coming principally from the state of Nuevo León and surrounding states (Coahuila, Tamaulipas and San Luis Potosí). HCV infection was diagnosed on the basis of clinical, biochemical and serological data. Demographic and histo-logical data and liver function test results were collected from all of the patients. The analysis of anti-HCV antibodies, HCV-RNA in plasma and HCV genotyping was performed according to the methods previously described.16 The research protocol was reviewed and approved by the hospital review boards. All participants signed informed consent forms at enrollment.
The control group included 100 healthy subjects who tested negative for HIV and HCV and other infectious diseases at the time of enrollment contemporaneously with case subjects. A peripheral blood sample was collected from each one. Forty-four were women and 56 were men. Mean age was 28 ± 11 years. The research protocol was reviewed and approved by institutional review boards. At enrollment, all participants signed the informed consent forms.
DNA isolation
Genomic deoxyribonucleic acid (DNA) was obtained from Ethylene-Diamine-Tetraacetic- Acid (EDTA)-preserved whole blood samples from patients and control subjects using the phenol-chloroform method as described before.17
Identification of TNF-α promoterpolymorphisms
Polymorphisms were detected by polymerase chain reaction (PCR) followed by the Restriction Fragment Length Polymorphism (RFLP) method. For polymorphism of TNF-a promoter at position -238 we used the following primers; sense primer 5'-AGACCCCCCTCGGAACC-3' and antisense primer 5'-ATCTGGAGGAAGCGGTAGTG-3'.18 while for -308 TNF-alpha polymorphism we used the sense primer 5'-CAATAGGTTTTGAGGGCCAT-3' (modified from Day et al. 1998).18 and the antisense primer described above. These primers containing a single base-pair mismatch adjacent to the polymorphism site to introduce a restriction site for the Msp I and Nco I enzymes (New England BioLabs, Beverly, MA, USA) into the wild-type nucleotide sequences after amplification for detecting the -238 and -308 polymorphisms, respectively.11,19 The amplified products of 151 bp and 223 bp were digested with Msp I and Nco I restriction enzymes and analyzed using 2% agarose gel.
Statistical Analysis
Descriptive statistics was used (mean and standard deviation). Hardy-Weinberg equilibrium was evaluated for these two polymorphisms within each group by the X2 test. Statistical significance was defined as P < 0.05. Data analysis was performed using the Epi-Info 2000 software version 3.3.2 and the SPSS version 13.0.
Results
We analyzed the frequency of -308 and -238 TNF-α polymorphisms in 48 patients with chronic HCV infection and 100 healthy non-related control subjects. The baseline characteristics of the patients with chronic HCV infection and healthy control subjects are given in Table 1. We also identified the HCV genotypes in these patients where the predominant genotype was 1 (79%), followed by genotype 2 (13%), 3 (6%) and 4 (2%) (Table 1). These results are similar to those reported previously for patients from northeastern Mexico and other regions of Mexico.16,20
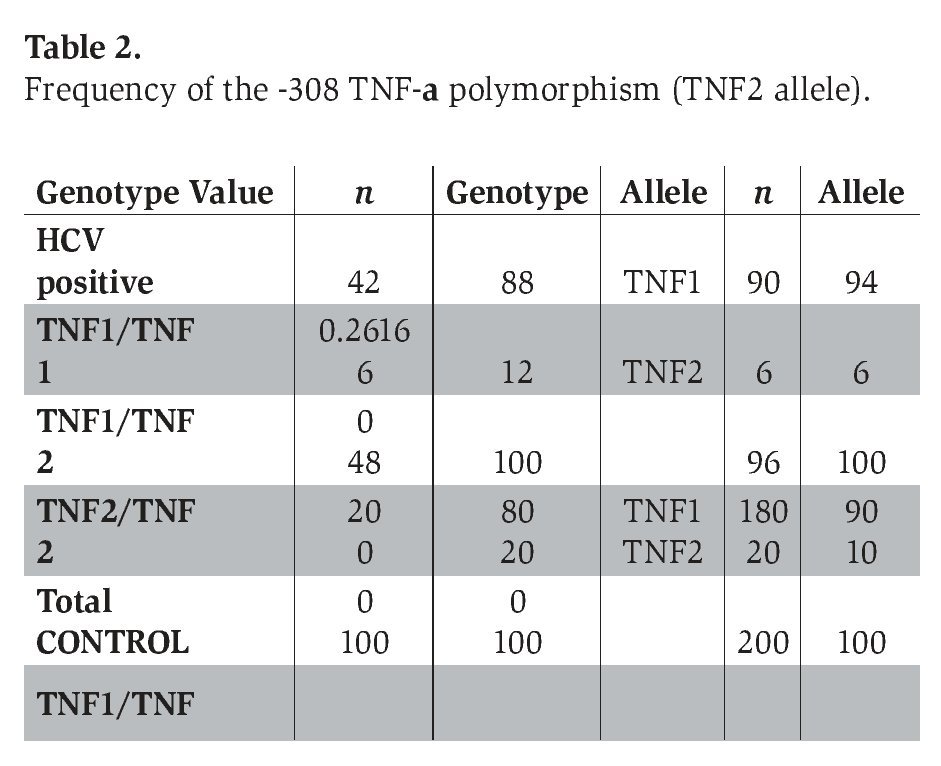
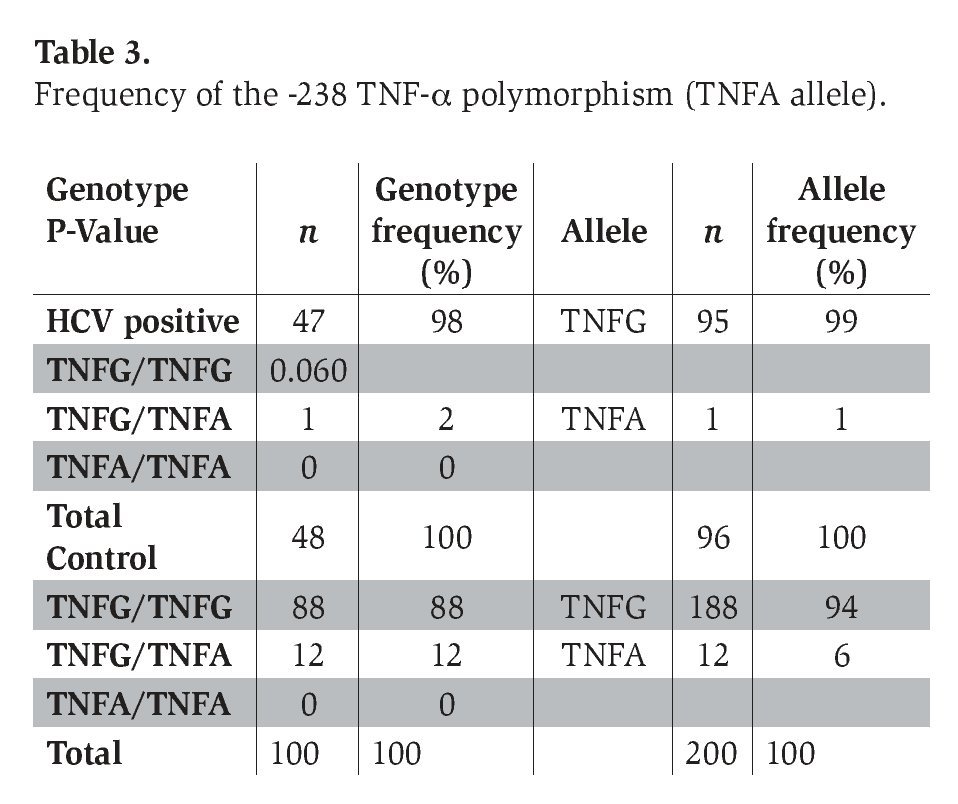
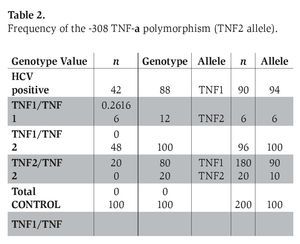
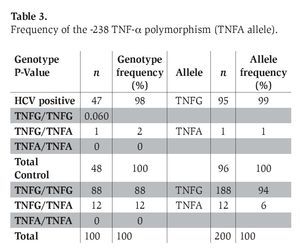
The genotype frequency of the -308 and -238 TNF-α polymorphism (with the prevalence of TNF2 and TNFA alleles) in patients with chronic hepatitis C infection and healthy control subjects is shown in Tables 2 and 3. The genotype frequencies of all of these polymorphisms did not deviate significantly from Hardy-Weinberg equilibrium in the two groups.
The prevalence of the -308 TNF-α polymorphism was 12% in patients with chronic hepatitis C and 20% in control subjects, respectively (P=0.2616) (Table 2).
DiscussionDai et al.13 reported that the TNF-α promoter geno-type at the position -308 appears to be associated with variability in severity of fibrosis and viral load in chronic HCV infection; nevertheless, we did not find a significant difference in the frequency of this polymorphism between these two groups. On the other hand, the prevalence of the -238 TNF-α polymorphism was 2% in HCV patients and 12% in control subjects (P=0.061). Hohler et al.12 reported that TNF-α gene promoter -238 polymorphisms were associated with chronic active hepatitis caused by HCV; however, no significant difference in the frequency of this polymorphism was observed between patients and the control group.
In addition, a previous study of the distribution of allele variants of the TNF-α gene carried out by German researchers failed to detect significant differences in the content of G-308A polymorphism genotypes in healthy subjects and patients infected with HCV (but they found allele associations with the disease for transition in locus -238).12 No appreciable differences in the incidence of TNF-α allele -308 were detected in a group of HCV infected patients belonging to different races.12,13 Previous studies found no association between TNF-α gene polymorphism in HCV patients (belonging to different races) and response to combined or monotherapy with IFN.12,13
Current reports indicate that TNF- effects on the course and outcome of HCV infection deserve further in vivo and in vitro investigations. The presence of a certain set of allele variants of cytokine genes, for example TNF- gene located in promoter regions, and hence, determining the level of spontaneous and inducible IL production by cells can be essential for the outcome of host contact with HCV and for the course of the infectious process and the efficiency of cytokine therapy.
In our study, the analysis of TNF-α promoter polymorphisms indicates that there was no significant difference between HCV infected patients and control subjects. These results are contrary to other reports showing that TNF-α promoter polymorphisms are associated with chronic hepatitis C.12,13
In conclusion, our results indicate absence of relationship among these TNF-α polymorphisms and occurrence of hepatitis C, suggesting that other factors are involved in the progression of this disease. However, functional studies in a larger population characterizing TNF-α transcriptional activity in CHC patients carrying -308 and -238 TNF-a variant allele(s) may help to clarify the role of these promoter polymorphisms in susceptibility to HCV infection.
Acknowledgments
We are especially grateful to the medical and technical staff at the Liver Unit of the Internal Medicine Department at the University Hospital of UANL. This study was supported by a grant from CONACYT (SALUD-2003-C01-002) awarded to AM Rivas-Estilla.
Correspondence author: Ana María Rivas-Estilla, PhD.
Department of Biochemistry and Molecular Medicine, School of Medicine, Autonomous
University of Nuevo León, Monterrey, NL, Mexico.
Phone: (52-81) 8329-4174; Fax: (52-81) 8333-7747;
E mail: amrivas1@yahoo.ca