Helicobacter pylori (H. pylori) is associated with gastroduodenal diseases. Virulence of clinical isolates is related to clinical outcome. Moreover, with microdiversity studies in clinical isolates from a single patient, but from a different origin (antrum or corpus), it is possible to demonstrate that there are simultaneous mixed infections.
AimsTo genotype H. pylori strains with multiplex PCR, according to their clinical virulence, and in this manner know the frequency of each genotype and relate it to clinical outcome in order to prevent the development of severe diseases.
MethodsA total of 210 patients with gastric alterations were studied. Virulence classification of H. pylori strains was carried out with multiplex PCR and 127 strains were identified as H. pylori by PCR (glmM and cagE). Genotype and clinical outcome were evaluated with the Fisher's exact test. In addition, RAPD-PCR was performed as a fingerprinting method to analyze mixed infections.
ResultsThe cagA, vacAs1, and vacAm1 genes were detected in all the clinical isolates. Strains were classified as: type i, 40.15% (51/127); type ii, 22.04% (28/127); and type iii, 28.4% (36/127), but two new different genotypes were also detected: (1) babA2+, cagA+, vacAs1+, 6.29% (8/127) and (2) babA2+, cagA-, vacAs2/m2+, 3.14% (4/127). The cagE gene was detected in type i strains.
ConclusionsThe Fisher's exact test did not support a significant association between clinical outcome and genotype. The main circulating genotypes in the Mexican population studied were: cagA+, vacAs1, and vacAm1. Multiplex PCR can be used as a screening test for H. pylori strains. Furthermore, the cagE gene is a good marker for identifying cag-PAI positive strains.
La infección por Helicobacter pylori se asocia con gastritis. La variabilidad genética de las cepas tiene importancia clínica. Los estudios de microdiversidad en cepas aisladas de diferente sitio anatómico en un mismo paciente revelan la posibilidad de recuperar 2 o más cepas diferentes.
ObjetivosGenotipificar por PCR multiplex según la virulencia de aislados clínicos de H. pylori para determinar la frecuencia de cada genotipo y relacionarlo con la evolución clínica con la finalidad de prevenir el desarrollo de enfermedades graves.
MétodosSe estudió a 210 pacientes con alteraciones gastroduodenales; 127 cepas fueron identificadas por PCR (glmM y cagE) y después clasificadas según su virulencia por PCR multiplex. Se hizo la prueba de Fisher para evaluar la relación entre genotipo y resultado clínico. La técnica de RAPD-PCR se empleó como método de huella genética y para analizar la presencia de infecciones mixtas.
ResultadoscagA, vacAs1 y vacAm1 estuvieron presentes en todos los aislados clínicos. Las cepas se clasificaron como: tipo i, 40.15%(51/127); tipo ii, 22.04% (28/127), y triples positivas, 28.4% (36/127); se detectaron 2 genotipos nuevos: 1) babA2+, cagA+, vacAs1+, 6.29% (8/27), y 2) babA2+, cagA–, vacAs2/m2+, 3.14% (4/127). cagE se detectó en las cepas tipo ii.
ConclusionesLa prueba de Fisher no mostró una asociación significativa entre el resultado clínico y el genotipo en la población estudiada. Los genotipos circulantes en la población mexicana fueron cagA+, vacAs1, vacAm1. La PCR multiplex puede usarse para genotipificar rápidamente las cepas de H. pylori. cagE es un buen marcador para identificar cepas cag-PAI+.
Helicobacter pylori (H. pylori) is a Gram-negative, spiral-shaped, microaerophilic bacterium associated with gastritis and peptic ulcer disease (PUD) and is considered a risk factor for gastric cancer.1–2 Half of the world population is infected with this bacterium, but only 10% of infected people will develop clinical outcomes. Infections with H. pylori may induce chronic gastritis, PUD, and gastric adenocarcinoma.3 There is evidence showing that the genetic variability of H. pylori strains has clinical importance. Three major virulence factors are used as epidemiological markers and to determine clinical outcome: the genes vacA (vacuolating toxin), cagA (cytotoxin-associated gene), and babA (BabA adhesin).4
The vacA gene, encoding the vacuolating toxin (VacA), is present in almost all strains. Significant polymorphism within vacA can be found in the middle region (m), with 2 alleles: m1 or m2 as well as in the signal sequence region (s) with 3: s1a, s1b, or s2. The possible genotypes are: s1bm1, s1am1, s1am2, s1bm2, s2m1, and s2m2, although the s2m1 genotype is reported to be rare.5 The s1am1 genotype, rather than m2, is associated with PUD.3 A third polymorphic determinant of vacuolating activity has been described and is called the intermediate (i) region. There are 2 i-region subtypes: i1 and i2 and variation among populations has been observed. Moreover, studies have shown a strong relation between the i1 allele and the production of CagA and the presence of the s1 type allele in various H. pylori strains isolated in individuals from several countries. This association could indicate that the intermediate region plays a role in more severe outcomes of chronic H. pylori infection.6 CagA is associated with an increased risk for gastric cancer. It is a protein that is injected into the interior of the cell through a type IV secretion system and is phosphorylated in the cell interior, inducing morphologic and probably cell proliferation changes, which favor the development of gastritis and gastric cancer.7 CagA is part of the cag pathogenicity island (cag-PAI) which also contains the cagE gene that has been associated with PUD in children and in adults with a severe clinical outcome, especially in the developed countries.8–9 BabA mediates the adherence of H. pylori to the human Lewis-b blood group antigen on gastric epithelial cells. It has a silent babA1 gene (unexpressed) and a babA2 gene that is expressed. The sequence of these genes differs by a deletion of 10 base pairs in the signal peptide sequence of babA1. A number of studies have suggested a relation between babA2-positive strains with an increase of higher H. pylori density, cellular mucosal inflammation, and a risk for developing severe clinical outcomes.10–12
Strains have been previously classified as type I (cagA+, vacA+) and type II (cagA-vacA-, according to the presence/absence of the genes. Type I strains are considered to be more virulent than type II strains.13 There is a third type of strain, called the “triple-positive strain”, with the presence of the babA+, vacA+ and cagA+ genes. These triple-positive strains have been associated with PUD and gastric adenocarcinoma.14
Fingerprinting studies have shown that independent H. pylori strains are distinguished from one another through random amplified polymorphic DNA (RAPD). Microdiversity studies in clinical isolates from different anatomic sites confirm the possibility of recovering 2 or more different strains from the same patient. Spontaneous mutations after serial passages in gnotobiotic piglets revealed that H. pylori might undergo host-specific adaptation allowing interstrain gene transfer to induce reorganization of its genome in order to adapt to a new environment.15-17
Recombinant strains could also emerge with different combinations of parental genotypes in mixed infections. Various DNA fingerprinting methods applied to several H. pylori isolates taken from the same patient have shown that all the different strains may be derived from an ancestral parent strain, but have undergone independent genomic changes, a phenomenon known as “microevolution”. Thus a single patient is able to carry two or more strains with a different genotype.17-18
In Mexico, there are studies that relate the presence of the microorganism with clinical outcome, gastric cancer, as well as with the presence of mixed infections, obtaining different genotypes in a single patient.19 A recent study reported the presence of 2 different alleles of the gene iceA in the same strain.20 The aim of our study was to genotype clinical H. pylori strains through multiplex PCR, according to their virulence classification, thus determining the frequency of each genotype and relating it to clinical outcome in order to prevent the development of severe diseases such as gastric cancer or PUD.
MethodsSubjects and clinical samples. A total of 210 patients with gastric alterations that underwent upper gastrointestinal endoscopy at the Hospital General “La Raza” of the Instituto Mexicano del Seguro Social (IMSS) in Mexico City, were evaluated. The age range of the pediatric patients in whom H. pylori was isolated was from 4 to 17 years. The only adult patient in whom H. pylori was isolated was 62 years old. Of those patients, 11 out of the 18 were male and 7 out of the 18 were female. The prevailing diagnoses were: gastroesophageal reflux disease, acid peptic disease, and recurrent abdominal pain.
Within the time frame of January 2003 to January 2005, 210 biopsies were obtained from each patient from the gastric corpus and were evaluated for the presence of H. pylori by the rapid urease test (RUT) and culture. RUT produced 30 (14.3%) positive biopsies from the gastric corpus. H. pylori was recovered by culture in 18 patients: 17 children and one adult. Written informed consent for participation in this study was obtained from each patient before the study. The Ethics Committee of the Hospital General approved the study protocol in advance.
H. pylori culture and subcloning. H. pylori was cultured from the biopsies. The specimens were homogenized in a sterile tissue grinder with a diluent (Phosphate Buffer Saline, pH 7.5). Homogenates were plated onto Casman agar base plates (Difco®) supplemented with 10% defibrinated sheep blood and Skirrow's selective supplement (vancomycin, trimethoprim, polymyxin B, OXOID®) and were incubated at 37°C with a supply of 5% CO2, 85%NO2, and 5% O2 for 4 days. Typical colonies were identified as H. pylori by morphology, through Gram staining (Gram-negative spiral or curved rods) and biochemical testing (positive urease, oxidase, and catalase tests). H. pylori ATCC 43504 was cultured as mentioned above and used as the positive control. A total of 18 clinical strains were isolated (recovery percentage, 13.3%) and they were each referred to as “pooled bacterial culture”. During the primary isolation process, 4-8 clinical isolates were separately picked from each plate and then propagated onto individual plates. Each one of these strains obtained was referred to as an “individual colony” or clinical isolate (n=127). The clinical isolates were maintained in cryotubes at −70°C in Hanks’ solution supplemented with 20% glycerol (Table 1).16,21
H. pylori isolates and subclones obtained from Mexican patients.
| Strain identification | Subject | Age (years) | Clinical isolates | RAPD subclones | Genotype | Diagnosis |
|---|---|---|---|---|---|---|
| 73 | C | 10 | 8 | 2 | babA2-cagA+vacAs1m1+ | AG |
| 126 | C | 14 | 8 | 1 | babA2+cagA- vacAs2m2+ | GU |
| B075 | C | 7 | 10 | 3 | babA2-cagA+vacAs1m1+ | NG |
| 78 | C | 14 | 8 | 2 | babA2-cagA+vacAs1m1+ | DU |
| 94 | C | 11 | 8 | 1 | babA2-cagA-, vacAs2m2+ | NG |
| 144 | C | 17 | 8 | 1 | babA2-cagA+vacAs1m1+ | NG |
| 175 | C | 16 | 6 | 1 | babA2+cagA+ vacAs1m1+ | MG |
| 39 | C | 11 | 10 | 1 | babA2-cagA+vacAs1m1+ | MG |
| B046 | C | 9 | 8 | 2 | babA2+cagA+ vacAs1m1+ | MG |
| B119 | C | 17 | 7 | 3 | babA2+cagA-vacAs1m1+ | GU |
| 58 | C | 8 | 6 | 1 | babA2-cagA-vacAs2m2+ | NG |
| 62 | C | 7 | 4 | 2 | babA2+cagA+vacAs1m1+ | MG |
| 29 | C | 5 | 4 | 2 | babA2+cagA+vacAs1+ | NG |
| 36 | C | 17 | 6 | 3 | babA2-cagA-vacAs2m2+ | NG |
| 63 | C | 7 | 7 | 1 | babA2+cagA+ vacAs1m1+ | NG |
| 163 | C | 15 | 7 | 2 | babA2-cagA+vacAs1m1+ | NG |
| 110 | C | 5 | 4 | 1 | babA2+cagA+ vacAs1m1+ | NG |
| 54 | Ad | 60 | 8 | 2 | babA2-cagA-vacAs2m2+ | MG |
Ad: adult; AG: acute gastritis; C: Child; Clones: subcloning strains; DU: duodenal ulcer; GU: gastric ulcer; MG: mild gastritis; NG: nodular gastritis; RAPD-PCR: different RAPD-PCR profile.
DNA extraction. DNA was extracted from the reference strain and from the clinical isolates of the colonies and collected in microcentrifuge tubes containing 100μL of PBS (phosphate-buffered saline, pH 7.5). Suspensions were vigorously vortexed for 2min and boiled in a water bath for 15min. The guanidine thiocyanate method for DNA extraction was used. The DNA pellet was suspended in TE (Tris-EDTA, pH 8.0) and frozen at -70°C until use.
Detection of the glmM gene. The presence of the glmM gene was used to monitor the purity of the DNA samples and to validate all subsequent analyses. Genomic DNA of the bacterial samples was amplified with the synthetic oligonucleotide primers, GLMM and GLMMR1 (Table 2), as previously described.22
Primers used for the amplification of vacA, cagA, babA2, cagE, and glmM alleles by conventional and multiplex PCR.
| Gene | Primer | Primer sequence 5’-3’ | Amplicon bp | Reference |
|---|---|---|---|---|
| vacAm1/m2a | VAGF | CAATCTGTCCAATCAAGCGAG | 570/645 | (Atherton et al., 1994) |
| VAGR | GCGTCAAATAATTCCAAGG | |||
| vacAs1/s2a | VAIF | ATGGAAAATACAACAAACACAC | 259/286 | (Atherton et al., 1994) |
| VAIR | CTGCTTGAATGCGCCAAAC | |||
| cagAa | F1 | GATAACAGGCAAGCTTTTTGAGG | 340 | (Tummuru et al., 1993) |
| R1 | CTGCAAAAGAATGTTTGGCAG | |||
| babA2a | babA2F | AATCCAAAAAGGAGAAAAAGTATGAAA | 812 | (Mizushima et al., 2001) |
| babA2R | TGTTAGTGATTTCGGTGTAGGAC | |||
| cagEb | F1 | GCGATTGTTATTGTGCTTGTAG | 329 | (Tsuneo et al., 2001) |
| R1 | GAAGTGGTTAAAAATCAATGCCCC | |||
| glmMb | GLMMF | GGA TAA GCT TTT AGG GGT GTT AGG GG | 140 | (Córdova et al., 2011) |
| GLMMR | GCA TTC ACA AAC TTA TCC CCA ATC |
RAPD-PCR (Randomly Amplified Polymorphic DNA-PCR) fingerprinting profiles. RAPD-PCR was performed to evaluate the genetic variability among the strains. The reaction was carried out in 25μL containing: 65 ng of H. pylori DNA, 0.8μM of the primers 1254, 1281, 1283 and 1247, 0.4mM of each dNTP, 1.5 U of TaqDNA polymerase (Invitrogen®), and 3mM of MgCl2. The cycling program used was 4 cycles of 94 oC, 5min; 36 oC, 5min; and 72 oC, 5min, and 30 cycles of 94 oC, 1min; 36 oC, 1min; and 72 oC, 2min, and then 72 oC, 10min in a Perkin-Elmer®, as previously described.22-23 RAPD-PCR bands were scored as present or absent and a binary matrix was assembled. The bands were assigned according to the molecular markers (1Kb, 100pb). A dendrogram was constructed with NTSYSpc-2.02j by the unweighted pair group method with the arithmetic mean (UPGMA) and a simple matching coefficient (SMC).24-25 Finally, the cophenetic correlation coefficient (CCrC) was obtained using the Mantel t-test and the best-cut test.26
Detection of the vacA, cagA, and babA2 genes by multiplex PCR. Multiplex PCR was carried out for genotyping the virulence genes (vacA, cagA, babA2). Genomic DNA of Proteus mirabilis was used as the negative control and H2O as the control in the reaction. The primers used for PCR are shown in table 2. Amplification was performed in a 25μL reaction volume containing 10 pM of babA2 primer and 5 pM of cagA and vacA primers, 10X Buffer solution (Invitrogen®), 1.5mM MgCl2, 25mM dNTPs (Invitrogen®), 80 ng DNA template, and 1 U Taq DNA polymerase (Invitrogen®). The cycling was performed in a Perkin Elmer® thermocycler with an initial denaturing step at 94 oC for 5min, followed by 35 cycles of 94 oC for 30 s, 57 oC for 45 s, and 72o C for 1min. Amplicons were distinguishable by size after separation through electrophoresis on a 1.5% agarose and revealed with ethidium bromide (0.5μg/ml).
The H. pylori ATCC 43504 amplicons obtained by multiplex PCR were purified with a Marligen kit (Bioscience®) according to the manufacturer's instructions and sequenced at the “Unidad de Síntesis y Secuenciación” IBT, UNAM at Cuernavaca, Morelos, for validating multiplex PCR. The sequences were compared with the sequences reported in the medical literature published in the NCBI database by BLAST analysis. Additionally, Southern-Blot and Dot-Blot were performed to compare and validate the results obtained from PCR.
Detection of the cagE gene. Amplification of the cagE gene was carried out as a cag-PAI marker, instead of the cagA gene. The primers used are shown in table 2. Amplification was performed in 25μL reaction volume containing: 10 pM of each primer, 10 X Buffer solution (Invitrogen®), 1.5mM MgCl2 (Invitrogen®), 25mM dNTPs (Invitrogen®), 80 ng DNA, and 1 U Taq DNA polymerase (Invitrogen®). The PCR protocol included an initial denaturing cycle at 94 oC for 5min, followed by 35 cycles of 94 oC for 60 s, 56 oC for 60 s, and 72 oC for 1min.
Data analyses, univariate analyses. A matrix of genotypes for all the genes encoded as present (1) or absent (0) was carried out. The percentage of positive alleles (present) for each gene was calculated from the data of all the strains. To evaluate whether the detection of the virulence genes (babA2, cagA, vacAs1m1) varied between conventional and multiplex PCR, the percentage of true and false positives was obtained using a chi-square test. To assess whether the presence or absence of the alleles (vacAs1, vacAs2, vacAm1, vacAm2, cagA, babA2) was associated with clinical outcome (acute gastritis, duodenal ulcer, gastric ulcer, nodular gastritis, and mild gastritis) a Fisher's exact test was done for each gene using a 2 X 2 contingency table in relation to the number of strains, with or without an allele, and clinical outcome.27
ResultsRAPD-PCR. The RAPD-PCR profile revealed 3 to 17 bands for each strain analyzed. The clones isolated from each patient are mentioned in table 1. RAPD-PCR detected the presence of multiple H. pylori strains, each different from the other, in a single person. Four primers produced genetic markers and reproducible RAPD profiles (virulence genotyping was performed for just 127 strains) among the strains studied. RAPD analysis grouped strains together based on clinical disease. The majority of the children were colonized by a single strain. The data suggest that, even though the H. pylori population consisted of very closely related bacteria, it was not completely homogeneous; genetic variants of the same H. pylori strains may exist among isolates. Two populations of H. pylori with different RAPD patterns colonized 7 children. However, 3 children were colonized by clones more closely related to strains of independent individuals than to each other (Table 1, Figs. 1 and 2). The results of the study indicated that there was multiple strain colonization in Mexican children. The CCrC was 0.86801 (range 0.6-0.9), showing a significant representation of the genetic relationship among the isolates.
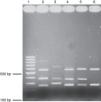
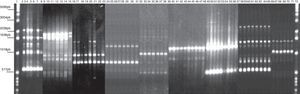
RAPD-PCR, 1281 oligonucleotide. 1Kb ladder molecular size marker, lane 1, control reagents, 2-8: Bx78 (78C2,7b,10,11,13,14 strains); 9-15:63 strain (63C1,2,4,5,6,7,22);16-23: B046 strain (B046C1, 5,9,10,11,13,14,15);24-31:29 strain (29C5,7,8,12,16,17,19,20);32-38:58 strain (58C1,2,4,8,9,13,15); 39-48:144 strain (144C1,2,4,6,8,20,21,22,23,25); 49-57:54 strain (54C1,2,3,5,6,7,8,11,13); 58-65:163 strain (163C1,2,5,10,11,30,31,34); 66-71:175 strain (175C2,4,7a,11,20,24); 72: 1Kb ladder molecular size marker.
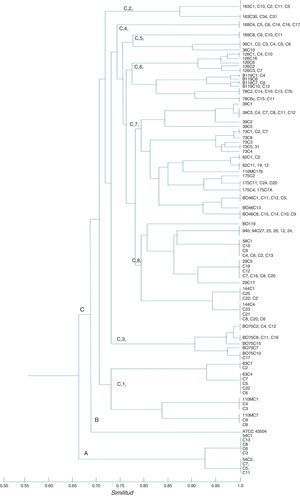
RAPD-PCR dendrogram of H. pylori strains. Three separated clusters are indicated with letters A, B, and C. Cluster A shows the adult strain (54); cluster B consists of the ATCC 43504 strain; cluster C consists of the pediatric Mexican strains (73, 126, B075, 78, 94, 144, 175, 39, B046, B119, 58, 62, 29, 36, 63, 163, 110, and 169); 169 strains were not genotyped. Group C shows 8 clusters. The CCCr was 0.86801. The past program and simple matching coefficient were used.
Multiplex PCR evaluation. One hundred and twenty-seven H. pylori clinical strains obtained from a pediatric population were characterized by conventional and multiplex PCR (Fig. 3) and the Southern blot and Dot blot results were correlated. The detection capacity of the Dot blot was: cagA (100pg/μl), vacAs1 (100 pg/μl), vacAm1 (100 pg/μl), and babA2 (1 pg/μl). Both methods detected the same number of true positive and/or negative strains for vacAs1m1, vacAs2m2, and cagA. However, the babA2 gene had a statistically significant difference (chi-square) (χ2) (Table 3).
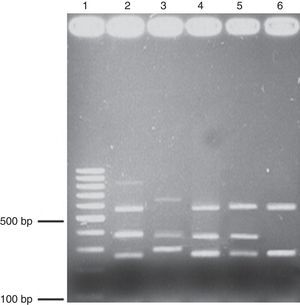
Multiplex PCR. Agarose (1.7%) gel electrophoresis showing the amplicons obtained through multiplex PCR. Lane 1, 100-bp ladder; lane 2, positive control triple virulence strains, lanes 3-6, clinical isolates. babA2 (812bp); vacAs1(259bp); vaAs2(286bp); vacAm1(570bp); vacAm2(645bp); cagA(340bp). The genomic DNA of the H. pylori ATCC 43504 strain was used as a positive control.
Comparison of virulence gene detection between conventional and multiplex PCR.
| Gene/allele | Value | Conventional | Multiplex | χ2 | p |
|---|---|---|---|---|---|
| babA2 | P | 48 | 15 | 82.323 | 1.16x1019 |
| N | 79 | 112 | |||
| cagA | P | 84 | 87 | 0.32845 | 0.56657 |
| N | 43 | 40 | |||
| vacAs1 | P | 91 | 91 | 0 | n.s |
| N | 36 | 36 | |||
| vacAs2 | P | 36 | 36 | 0 | n.s |
| N | 91 | 91 | |||
| vacAm1 | P | 87 | 87 | 0 | n.s |
| N | 40 | 40 | |||
| vacAm2 | P | 36 | 36 | 0 | n.s |
| N | 91 | 91 |
N: negative value; n.s.: not significant; P: positive value; χ2: chi-square statistic.
We obtained false negative values when the results were compared by conventional PCR.
H. pylori strains: classification and genotyping. A high frequency of the cagA, vacAs1, and vacAm1 genes was detected in all the clinical isolates. The results of the genes are shown in Table 4. The strains genotyped by multiplex PCR were classified as type I, type II, and triple-positives, as classified by Atherton5 and Gerhard.14 In addition, we found 2 genotypes different from the type I, II, and triple-positive strains, which were described as “non-typeable”. In one strain (B119) and its clinical isolates (n=7), the cagA gene was negative using conventional or multiplex PCR, but detectable when the Dot Blot was used, so all of them were classified as triple-positive strains. The vacAs1 and vacAm1 genotypes were present in almost all of the clinical outcomes and the cagA genes were more frequently associated with gastritis and gastric ulcer disease. The babA2 gene was present in patients with gastric ulcer and mild gastritis, but the Fisher's exact test did not support a significant association between clinical outcome and the genes studied (Table 5). The babA2 gene was detected in 48 clinical isolates and it was distributed mainly in triple-positive strains (n=29). The babA2 gene was also detected in the “non-typeable” strains mentioned above as follows: babA2+, cagA+, vacAs1+ (n=8), babA2+, cagA-, vacAs2/m2+ (n=4). Each patient was infected with only one type of strain (type I, II, III or with the “non-typeable” strain) (Table 6). The cagE gene was amplified by PCR. Amplification of the cagE gene has been used for evaluating the presence or absence of cag-PAI since the absence of the cagA gene in H. pylori strains does not mean that the cag-PAI is completely absent. The cagE gene was detected in all type II strains from patients with ulcer disease (100%), thus the cagE gene can be a useful and better marker than the cagA gene for detecting the presence of the whole cag-PAI pathogenicity island.
Percentage and total number of H. pylori pediatric strain virulence markers babA2, cagA, and vacAs1m1.
| Gene/Allele | Total number | 95% confidence limits | Percentage |
|---|---|---|---|
| vacAm11 | 89 | 61.3–77.9 | 68.5 |
| vacAm2 | 34 | 19.3–35.4 | 28.34 |
| vacAm- | 4 | 0.9–7.9 | 3.16 |
| vacAs1 | 93 | 64.6–80.7 | 71.63 |
| vacAs2 | 34 | 19.3–35.4 | 28.34 |
| cagA+ | 84 | 57.2–7.3 | 66.14 |
| cagA- | 43 | 25.7–42.8 | 33.86 |
| babA2+ | 48 | 29.3–46.8 | 37.79 |
| babA2- | 79 | 53.2–70.7 | 62.21 |
High percentages. EpiInfoTM 2000 program.
babA2: gene that encodes BabA adhesin targeting Lewisb (α-1, 3/4-difucosylated) blood group antigens; cagA: gene encoding a highly immunogenic protein CagA;
vacA: vacuolating cytotoxin gene.
Highlighted in black: the highest percentages of each gene or allele.
Association between gene virulence and clinical outcome.
| Gene % | AG n=1/18 | DU n=1/18 | GU n=2/18 | NG n=9/18 | MG n=5/18 | Total number |
|---|---|---|---|---|---|---|
| vacAm1 | 1 (100)* | 1 (100)* | 1 (50)* | 5 (55.5)* | 4(80)* | 12 |
| vacAm2 | 0 (0)* | 0 (0)* | 1 (50)* | 3 (33.3)* | 1(20)* | 5 |
| vacAs1 | 1 (100)* | 1 (100)* | 1 (50)* | 6 (66.6)* | 4(80)* | 13 |
| vacAs2 | 0 (0)* | 0 (0)* | 1 (50)* | 3 (33.3)* | 1(20)* | 5 |
| cagA | 1 (100)* | 1 (100)* | 0 (0)* | 6 (66.6)* | 4(80)* | 12 |
| babA2 | 0 (0)* | 0 (0)* | 2 (100)* | 3 (33.3)* | 3(60)* | 8 |
AG: acute gastritis; DU: duodenal ulcer; GU: gastric ulcer; MG: mild gastritis; NG: nodular gastritis.
Classification of the H. pylori strain genotypes.
| Classification | Genotype | Total strains n=127 (%) | CO | Number of patients |
|---|---|---|---|---|
| I | babA2-, cagA+, vacAs1m1+ | 51 (40.15) | AG,NG,DU, MG | 6 |
| II | babA2-, cagA-, vacAs2m2+ | 28 (22.04) | NG,MG | 4 |
| III | babA2+, cagA+, vacAs1m1+ | 29 (22.83) | NG,MG | 5 |
| 1 | babA2+, cagA+, vacAs1+ | 8 (6.29) | NG | 1 |
| 2 | babA2+, cagA−, vacAs2m2+ | 4 (3.14) | GU | 1 |
| 3 | babA2+, cagA-,vacAs1m1+ | 7 (5.51) | GU | 1 |
AG: acute gastritis; CO: clinical outcome; DU: duodenal ulcer; GU: gastric ulcer; MG: mild gastritis; NG: nodular gastritis.
There are many advantages for employing a multiplex PCR. One example is when working with fastidious microorganisms or nosocomial ones, since this methodology increases the efficiency of detection and lowers the cost of the assays. PCR multiplex can be useful for genotyping clinical isolates obtained from biopsies or cultures and accurately diagnosing the presence of an infection due to H. pylori, but also for differentiating species of Helicobacter other than H. pylori.28-29 In our study, the sensitivity and specificity of multiplex PCR were excellent for all the genes. Multiplex PCR can be used as a discriminatory test to distinguish type I strains, which are more virulent and associated with severe clinical outcomes. Some authors have stated that there can be a relationship between strain genotype and clinical outcome, enabling the prediction of the latter.30 Other authors have reported that triple-positive strains (babA2+, cagA+, vacAs1m1+) were associated with duodenal ulcer and adenocarcinoma.14 In adult populations, triple-positive strains are linked to severe diseases. However, we did not find any relationship between strain genotype and clinical outcome in the pediatric population studied. According to our results, type II clinical isolates were more related to non-atrophic gastritis.
The clinical isolates were classified as type I (n=51), type II (n=28), and triple-positive (n=29). We used the Dot blot assay to detect the cagA gene in the B119 strain (n=7) because it was not possible to detect the gene by conventional PCR or multiplex PCR. Therefore, the number of triple-positive strains (n=36) increased. Nineteen clinical isolates could not be genotyped using the conventional classification (type I, II, or triple-positive), so we called them “non-typeable” strains: babA2+, cagA+, vacAs1+ (n=4) and babA2+, cagA−, vacAs2m2+ (n=8).
The variability of the vacA gene has been previously associated with geographic regions that include Colombia, the southeast region of Brazil, the United States, as well as in other Western populations, and strains with a vacAs1m1 genotype were considered higher cytotoxin producers, mainly associated with symptomatic children, patients with PUD, and patients with functional dyspepsia. However, in China, Japan, and South Korea, this genotype is not as predominant and there is no relationship.31 In the present study, the m1 allelic form of the vacA gene was present in all the clinical isolates and in patients with PUD (68.5%). Patients infected with strains having the s1 (71.63%) allelic form, as well as strains containing the vacAs2m2 and vacAs1m1 genotypes, were correlated with acute gastritis, mild gastritis, nodular gastritis, and duodenal ulcer. However, no conclusive relationship between strain genotype and clinical outcome was found. In our study, the cagA+ strains (66.14%) were isolated from patients with gastritis and peptic ulcer disease.
We detected type II strains (n=28) isolated from patients with nodular and mild gastritis. The amplification of the cagE gene was done to verify the presence of cag-PAI, since this gene is present in all type II strains. Given that the absence of cagA does not necessarily mean the lack, interruption, or partial loss of the cag-PAI, we amplified the cagE gene, using it as an optional marker to verify the lack of cag-PAI and finally to correlate the type II strains, cagE+, and severe clinical outcomes. In our population, the babA gene was detected at a low percentage (37.79%). Contrastingly, other South American studies have reported a frequency ranging from 46% to 82.3%.31 In our study, the babA gene was found mainly in patients with gastritis (n=6) and PUD (n=2), as has been reported in others countries.32-33
Type I strains (n=51) were isolated from patients with gastritis (n=5) and duodenal ulcer disease (n=1). Our results are similar to those reported in a western population, in which the prevalence of this genotype was 60-90%. However, the sample size must be increased to establish a relationship between strain genotype and clinical outcome.
The triple-positive strains (cagA, vacAs1m1, babA2) were isolated from patients with gastritis (n=5), but we could not establish a relationship between genotype and clinical outcome. Nevertheless, in several studies in Colombia, it was possible to correlate peptic ulcer disease and gastric carcinoma with these genotypes.14,31 In western countries, these types of strains are a risk factor, but no relationship with severe clinical outcomes has been found in Japanese populations. In the present study, all the clinical isolates were analyzed by RAPD-PCR, and in the same patient we observed some clinical isolates with different fingerprinting patterns, indicating that the patient presented with a mixed infection (2 strains with genetic differences derived from strain 73). All results from multiplex PCR of these different isolates from the same patient had the same genotype (babA2, cagA, vacAs1m1). Similar results were observed in the 39C5, B119C2, 29C17, 36C10, and 110M17 clinical isolates.
Strain 126 and its respective clones (126C1, 126C2, 126C3, 126C4, 126C6, 126C7, 126C10, 126C16) were isolated from a 14-year-old patient with gastric ulcer. In this strain, the same RAPD fingerprinting pattern with the same genotype (babA2+cagA-, vacAs2m2+) was observed. Therefore, the babA+gene in these strains could increase the risk for developing gastric ulcer.
In conclusion, multiplex PCR has been used as a rapid assay for detecting more than one virulence factor in a single reaction, and also as a method for diagnosing diseases, lowering the cost of the test for patients, and improving treatment. Our study proposes the use of multiplex PCR as a strategy for screening H. pylori clinical isolates and classifying them according to the virulence factors in a quick and easy manner, to prevent the development of severe diseases and to be used as a clinical monitoring tool for patients that have already been diagnosed. The results obtained in the present study show a variety of virulence genotypes in Mexico. The prevalent strain genotype circulating in a Mexican population was type I (babA2-, cagA+, vacAs1m1) and was present in all the clinical outcomes. The cagE gene was a better marker than cagA for detecting cag-PAI-positive strains.
Ethical responsibilitiesProtection of persons and animalsThe authors declare that the procedures followed conformed to the ethical standards of the responsible committee on human experimentation and were in accordance with the World Medical Association and the Declaration of Helsinki.
Data confidentialityThe authors declare that they have followed the protocols of their work center in relation to the publication of patient data.
Right to privacy and informed consentThe authors have obtained the informed consent of the patients and/or subjects referred to in the article. This document is in the possession of the corresponding author.
Financial disclosureFinancial support for this work was received from the SIP-IPN (20130644), CONACYT, SNI, EDI, and COFAA Institutions.
Conflict of interestThe authors declare that there was no conflict of interest.
The authors wish to thank the Instituto Mexicano del Seguro Social and Dr. Consuelo Ruelas-Vargas from the Department of Pediatric Endoscopies at the Hospital General “La Raza” for the help provided in the collection of specimens.
Please cite this article as: González-Vázquez R, Córdova-Espinoza MG, Escamilla-Gutiérrez A, Morales-Méndez I, Ochoa-Pérez SA, Armendáriz-Toledano F, et al. Frecuencia de genes de virulencia en infecciones mixtas con cepas de Helicobacter pylori de una población mexicana. Revista de Gastroenterología de México. 2016;81:11–20.