Helicobacter pylori (H. pylori) eradication treatment includes a proton pump inhibitor and two antibiotics: amoxicillin and clarithromycin. The goal of that treatment is to eradicate the infection in at least 90% of the patients. Failure to eradicate the infection can have multiple causes, among which is the presence of point mutations in the antimicrobial target genes.
ObjectiveTo characterize the mutations present in the pbp1a gene and their possible association with resistance to amoxicillin in vitro.
MethodologySusceptibility to amoxicillin was evaluated in 147 isolates of H. pylori from the Colombian municipality of Túquerres. PCR amplification and sequencing of the glycosyltransferase domain of the pbp1a gene were carried out on Túquerres isolates, and the association between mutations and resistance was evaluated.
ResultsA total of 5.4% (8/147) Túquerres isolates were resistant to amoxicillin in vitro. PCR amplification of the glycosyltransferase domain of the pbp1A gene was performed on 87.5% of the amoxicillin-resistant isolates in vitro, and in the DNA sequencing analysis, a total of 2 changes of amino acids from 3 DNA mutations that encoded the PBP1A-1 protein were observed.
ConclusionThe present study is the first report on pbp1a gene mutations in H. pylori isolates coming from a population in Túquerres. Mutations that have not been reported in previous studies were found.
El tratamiento para la erradicación de Helicobacter pylori (H. pylori) incluye un inhibidor de la bomba de protones y 2 antibióticos: amoxicilina y claritromicina; este tratamiento tiene como objetivo erradicar la infección en al menos el 90% de los pacientes. El fracaso del tratamiento de erradicación de la infección puede tener múltiples causas, entre ellas la presencia de mutaciones puntuales en los genes diana de los antimicrobianos.
ObjetivoCaracterizar las mutaciones presentes en el gen pbp1a y su posible asociación con la resistencia a la amoxicilina in vitro.
MetodologíaSe evaluó la susceptibilidad a amoxicilina en 147 aislados de H. pylori de Túquerres, se amplificó por PCR y secuenció los dominios transglicosilasa del gen pbp1a en aislados de Túquerres, y se evaluó la asociación entres las mutaciones y la resistencia.
ResultadosSe observó que el 5.4% (8/147) de aislados de Túquerres fueron resistentes a amoxicilina in vitro. La amplificación por PCR de los dominios transglicosilasa del gen pbp1a se realizó en el 87.5% de los aislados resistentes in vitro a amoxicilina, y en el análisis de la secuenciación del ADN se observó un total de 2 cambios de aminoácidos a partir de 3 mutaciones en el ADN que codifica para la proteína PBP1A-1.
ConclusiónEsta investigación constituye el primer reporte sobre las mutaciones en el gen pbp1a en aislados de H. pylori provenientes de las poblaciones de Túquerres; se encontraron mutaciones no reportadas en investigaciones previas.
Helicobacter pylori (H. pylori) is a Gram-negative, spiral-shaped, microaerophilic bacterium, with flagellates that enable its motility toward and adherence to the gastric epithelium.1 The bacterium is associated with gastritis and peptic ulcer disease and is considered a risk factor for gastric cancer.1
H. pylori has virulence factors that enable it to colonize the gastric mucosa and develop the disease, inducing a higher incidence and prevalence of its pathogenicity. The most important virulence factors are: urease, the flagellum, the cagA gene, the vacA cytotoxin, and the BabA protein.2
H. pylori colonizes the gastric mucosa of over 50% of the world population. It is generally acquired in childhood and persists for a lifetime. Notably, the clinical result of the infection is influenced by environmental factors, the genetic diversity of the strain, and the genetic diversity of the host.3
Regarding treatment, the first-line regimen for H. pylori eradication is the triple therapy that includes a proton pump inhibitor and 2 antibiotics, such as amoxicillin and clarithromycin, and its aim is to eradicate the infection in at least 90% of patients.4 Eradication treatment failure can be due to inadequate doses, a lack of treatment adherence, the presence of efflux pumps, and mutations in the genes targeted by the antimicrobials.
Antimicrobial resistance is a natural phenomenon that is accelerated by the selective pressure on microorganisms produced by exposure to antibiotics. As a result, conventional antibiotic treatments become inefficacious, infections persist, and the risk of their propagation increases.5
One of the most widely used antibiotics in eradication therapy is amoxicillin, a semi-synthetic derivative of ampicillin. It inhibits cell wall biosynthesis, binding to the penicillin-binding proteins (PBPs)6 in the cytoplasmic membrane. PBP1A has glycosyltransferase/transpeptidase activity, and its function is to elongate the peptidoglycan strands and form the cross-linking between the peptidoglycan peptides. Thus, its inhibition interferes with peptidoglycan synthesis and cell growth.7
The prevalence of H. pylori resistance to amoxicillin varies geographically, with values of 2.2%, 65.6%, 11.6%, and 0.5% for America, Africa, Asia, and Europe, respectively.8,9 Even though information on the prevalence of resistance in Colombia is limited, it has been reported at 7% in Bogotá,10 3.8% in the coffee-growing region,11 and 20.5% in Tumaco.12 None of those studies evaluated the pbp1a gene and its association with resistance to amoxicillin. Mutations in the pbp1a gene and their association with resistance to amoxicillin is unknown, with respect to H. pylori isolates from a population in Túquerres, located in the Colombian Andes; with the prevalence of H. pylori (> 80%) and high risk for the development of gastric cancer;13 as well as the possible association between the mutations in that gene and H. pylori eradication failure. Such information is useful for the development and implementation of methodologies for the rapid detection of resistance and the application of eradication regimens. Therefore, the aim of the present study was to characterize the mutations present in the pbp1a gene and their possible association with resistance to amoxicillin in vitro.
Materials and methodsAn observational study was conducted that included 149 samples of gastric mucosa taken from patients from the municipality of Túquerres, in the department of Nariño, Colombia, that had symptoms of dyspepsia, for the purpose of evaluating antimicrobial resistance to amoxicillin in vitro, through the agar dilution method. To achieve the proposed aims, gastric mucosa biopsy samples were taken, after which cultures were made and biochemical tests were carried out, to identify pure H. pylori colonies, confirmed through PCR molecular identification of the structural gene, ureA. Antimicrobial susceptibility to amoxicillin in vitro was then evaluated, and finally, the glycosyltransferase domains of the pbp1a gene were amplified and sequenced for the identification of point mutations. A detailed description of the methods utilized follows below.
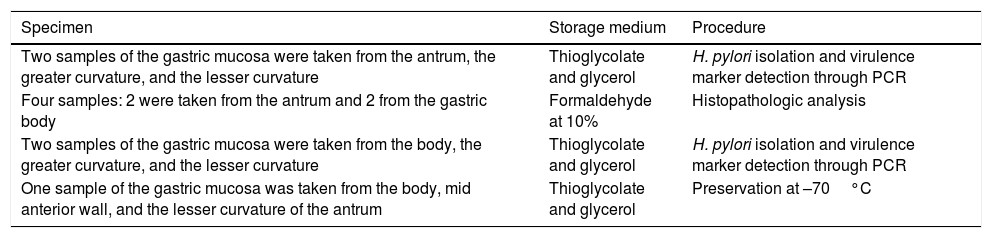
Gastric mucosa biopsyThe sample was selected from adults of both sexes that were referred to the medical service for clinical evaluation. Gastroduodenoscopy was ordered as part of their evaluation due to symptoms that included abdominal pain, gastroesophageal reflux, vomiting, respiratory symptoms, dyspepsia, and diarrhea, reported by the patients during the clinical consultation. Table 1 describes the biopsy protocol.
Endoscopic biopsy protocol.
| Specimen | Storage medium | Procedure |
|---|---|---|
| Two samples of the gastric mucosa were taken from the antrum, the greater curvature, and the lesser curvature | Thioglycolate and glycerol | H. pylori isolation and virulence marker detection through PCR |
| Four samples: 2 were taken from the antrum and 2 from the gastric body | Formaldehyde at 10% | Histopathologic analysis |
| Two samples of the gastric mucosa were taken from the body, the greater curvature, and the lesser curvature | Thioglycolate and glycerol | H. pylori isolation and virulence marker detection through PCR |
| One sample of the gastric mucosa was taken from the body, mid anterior wall, and the lesser curvature of the antrum | Thioglycolate and glycerol | Preservation at –70°C |
In accordance with the guidelines of the Colombian Department of Health, the samples were stored in media and then transported to the pathology laboratory of the Universidad del Valle, where the corresponding analyses were performed.
Gastric biopsy cultureThe initial culture, from which H. pylori from the gastric biopsies was isolated, was made by placing the biopsy sample in 200μl of sterile NaCl at 0.89% and macerating it in a sterile macerator. The resulting maceration was then taken with a disposable inoculating loop (Fisherbrand®, calibrated at 10μl/drop) and streaked in Columbia agar (Oxoid ®) with 10% defibrinated sheep blood plus Dent supplement (Oxoid®), containing vancomycin, cefsulodin, trimethoprim lactate, and amphotericin B. After the culture was made, the rest of the gastric mucosa maceration was preserved in a thioglycolate solution (Merck®) with 20% glycerol (Promega®) and stored at –70°C.
The Petri dishes were incubated in a CO2 incubator (Shel Lab®) at 37°C, with an atmosphere of 10% CO2 and 90% humidity. Growth was evaluated at 72h and then periodically, for 7 to 10 days, searching for small, translucent colonies in the shape of dew drops, consistent with the morphology of H. pylori. The quality of growth and fungal or bacterial contamination in the primary isolate were evaluated, as was the antibiogram.
Culturing the preserved samplesTo evaluate the susceptibility to amoxicillin in the 147 preserved and viable isolates of H. pylori from a Túquerres population, the bacterium had to be isolated again from the preserved samples. To do so, a fraction of the preserved maceration was taken (–70° C), utilizing a sterile lancet, and deposited in Columbia agar (Oxoid®), with 10% defibrinated sheep blood and no supplement. It was spread en masse and incubated at 37° C in a microaerophilic environment (10% CO2 and 90% humidity). Growth was evaluated at 72h and then periodically, for 7 to 10 days. The remaining content of the preserved maceration was stored at –70°C.
Purification of the Helicobacter pylori isolatesThe previously obtained colonies that had a phenotype consistent with H. pylori were streaked in triplicate, utilizing a sterile loop (10μL), in Columbia agar (Oxoid®), plus 10% defibrinated sheep blood, with the Dent supplement (Oxoid®), containing vancomycin, cefsulodin, trimethoprim lactate, amoxicillin, and amphotericin B. The Petri dishes were incubated (Shel Lab® incubator) at 37°C in a microaerophilic environment (10% CO2 and 90% humidity). Growth was initially evaluated at 48 to 72h, followed by periodic evaluations every 24h for 7 to 10 days. Phenotype identification tests and ureA gene amplification were performed on the resulting colonies.
Helicobacter pylori phenotype identificationSmall, punctate, translucent colonies, with well-defined edges were macroscopically searched for, and the following identification tests were carried out on them: the oxidase test, catalase test, urea test (host test), and Gram stain test. Lastly, for their correct identification, they were compared with the results presented in Bergey’s Manual of Systematic Bacteriology for the identification of H. pylori. In addition, molecular identification was performed through PCR amplification of ureA, the structural gene of H. pylori.
Evaluation of H. pylori isolate susceptibility to amoxicillin through the agar dilution methodAntimicrobial susceptibility was determined by applying the agar dilution method to H. pylori isolates from the patients, in which the cutoff point for considering a strain resistant to amoxicillin was defined, with a minimum inhibitory capacity (MIC) greater than or equal to 1.0μg/mL (Figueroa et al., 2012).
That was carried out on the 147 H. pylori isolates obtained from the initial Columbia agar culture. An inoculate of approximately 6×108 CFUs/mL (McFarland 2) was spread, in triplicate and by depletion, in Mueller Hinton (Merck®) agar, supplemented with 10% defibrinated sheep blood, at doubled concentrations (0.25, 0.5, 1, 2, and 4.0μg/mL) of amoxicillin, and incubated (Shel Lab® incubator) at 37° C in a microaerophilic environment (10% CO2 and 90% humidity). Bacterial growth was evaluated after 72h. The H. pylori ATCC 43504 was used as the quality control strain for monitoring the accuracy of the MIC of H. pylori, when using the agar dilution method.
DNA extractionFor bacterial DNA extraction, 1ml of the H. pylori isolates preserved (80% thioglycolate and 20% glycerol) at –70°C was taken and deposited in a 1.5ml Eppendorf tube. It was then centrifuged (Forma Scientific® refrigerated centrifuge) at 13,000rpm for 2min. The supernatant was discarded and 300μl of lysis buffer (Proteinase K 100μg/mL, SDS 10%, EDTA 0.5M and Tris-HCl pH 8) was added. The solution was incubated at 56°C for 18h, followed by 10min at 72°C. After incubation, 120μl of NaCl 5M was added and centrifuged at 13,000rpm for 5min. The supernatant was transferred, and the precipitate was discarded. A total of 840μl of absolute ethanol (Mallinckrodt®) was added and centrifuged at 13,000rpm at 4°C for 20min. The supernatant was discarded and 300μl of 70% ethanol was added and centrifuged at 13,000rpm for 5min, discarding the supernatant. The solution was dried for 2h, then resuspended in 100μl of TE buffer, and stored at –20°C. The quantity and purity of bacterial DNA was determined by an optic density reading of 260/280nm in a spectrophotometer (Gene Quant II® Pharmacia Biotech), according to the manufacturer’s instructions.
UreA gene amplificationThe molecular identification of H. pylori was performed through PCR amplification of the ureA gene. The reaction was carried out in a thermocycler (Swift MiniProTM, Esco), adding the following reagents to a 0.2ml tube: 1 X of PCR buffer (Promega® 5X Green Buffer), 1 μM of MgCl2 (Promega®), 0.25mM of dNTPs (Promega®), 50pmol/μl of each primer (sense 3'-AAGACATCACTATCAACG-5'/ antisense 5'-CCCGCTCGCAATGTCTAA-3’), 0.5 units of GoTaq DNA polymerase (Promega®); and 25ng of H. pylori genomic DNA at a final volume of 25μl. Amplification was carried out through an initial denaturation at 95°C/2min, followed by 35 cycles (95°C/1min, 54°C/1min, and 72°C/1min) and a final extension at 72°C/15min. The amplification products were obtained by electrophoresis at 80volts for 1h in a horizontal chamber (Spectroline Bi-O-Vision®). They were visualized through fluorescence in UV light (260/280nm) in a transilluminator (Spectroline Bi-O-Vision®), in 2% agarose gels (Sigma®) stained with 1μl of ethidium bromide (0.5μg/mL). To determine the amplified products of all the genes, a 100 bp molecular weight marker (Fermentas ®) was used. The size of the amplicon corresponded to approximately 167 bp (expected fragment through an in silico analysis, with respect to the migration distance of the base pair marker).
Amplification of the pbp1a gene glycosyltransferase domains in Helicobacter pyloriPCR amplification of the glycosyltransferase domains (pbp1a) was carried out using a thermocycler (Swift MiniProTM, Esco) and the following reagents were added to a 0.2ml tube: 1 X PCR buffer (Promega 5X Green Buffer®), MgCl2 1mM (Promega®), 10% DMSO, dNTPs 0.288mM (Promega®), 50pmol/μl of each pbp1a-1 primer [R-GCCATTCTTATCGCTCAAGTT and F-TCTCGTGTGAGCACCATG], 0.5 units of GoTaq DNA polymerase (Promega®); and 25ng of H. pylori genomic DNA, at a final volume of 50μl. The amplification thermal cycle consisted of an initial denaturation at 95°C/2min, followed by 35 cycles (95°C/1min, 54°C/1min, 53°C/1min) for pbp1a-1 (glycosyltransferase domain), then at 72°C/1min, and a final extension at 72°C/5min.
The PCR amplification products were obtained through electrophoresis at 80volts for 1h in a horizontal chamber (Spectroline Bi-O-Vision®). They were viewed through fluorescence in UV light (260/280nm) in a transilluminator (Spectroline Bi-O-Vision®) in 2% agarose gel (Sigma®), stained with 1μl of ethidium bromide (0.5μg/mL). To determine the size of the amplified products of all the genes, a 100 bp molecular weight marker (Fermentas ®) was used. The amplicon size was 340 bp for the pbp1a-1 glycosyltransferase domain. The primers were designed using the sequence for the GenBank pbp1a gene code: AE000511.1 in the Primer3 V 0.4.0 software.14
Mutation sequencing and identificationThe purified amplified products of the pbp1a gene were sequenced in both senses (sense and antisense), employing the genetic analyzer (ABI 3130 Applied Biosystem®) and the Big Dye Terminator (Applied Biosystem®) methodology, according to the standardized conditions at the Human Molecular Genetics laboratory of the Universidad de Valle. BioEdit v 7.1.11® software was utilized in the edition, alignment, and translation of the sequences. The changes in the sequences were collated through local alignment, with GenBank accession: AE000511.1, for the pbp1a gene of H. pylori strain 26695.15
The alignment was exported in the FASTA format and opened in the notepad. The information was copied and pasted onto a book in Excel, adding the logical formula (Datum X=Datum Y)*1, which presented values of 0 for different data between the aligned sequences and 1 for equal data. The changes were determined for each sample. For the pbp1a samples, the DNA was additionally translated to protein in the BioEdit software and the same analysis was performed to determine the changes.
Statistical analysisThe frequency and proportion of the resistant isolates from a Túquerres population were calculated. The performance of the amplification and sequencing of the pbp1a gene fragments was evaluated through descriptive analyses. The frequency of the mutations in the genes was calculated. Only descriptive analyses were carried out for the mutation variables in the pbp1a gene due to the low number of samples, upon comparing the resistance in vivo with the resistance in vitro (between 0 and 3 data).
Ethical considerationsThe present study was approved by the Comité Institucional de Revisión de Ética Humana (CIREH) of the Faculty of Health of the Universidad del Valle, regulated by Resolution 008430 from October 4/1993, issued by the Colombian Department of Health.
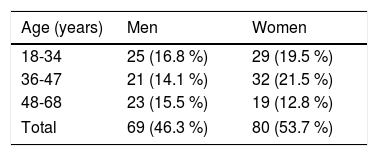
ResultsOf the 149 participants, 53.7% (80/149) were females, 29 of whom were between 18-34 years of age. Of the males, 25 were between 18-34 years of age (Table 2).
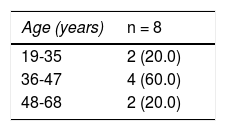
The bacterium was isolated from 147 of the 149 participants. Upon evaluating the antimicrobial resistance to amoxicillin in vitro in the H. pylori isolates, through the agar dilution method, 139/147 (94.6%) of the isolates were susceptible and 8/147 (5.4%) were resistant to amoxicillin in vitro (Table 3). Distribution by sex was 50% in the resistant isolates and they were obtained from patients between 19-35, 36-47, and 48-65 years of age (Table 4).
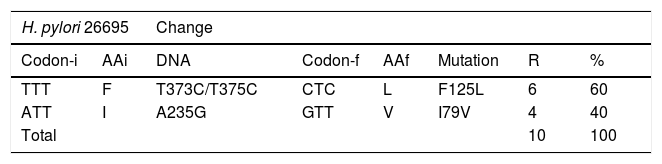
PCR amplification of the ureA gene was positive for the 147 isolates obtained and PCR amplification of the glycosyltransferase domains (PBP1A) of the pbp1a gene of H. pylori was performed on 87.5% of the isolates resistant to amoxicillin in vitro. In the DNA sequencing analysis of the 8 resistant isolates, there was a total of 2 changes of amino acids. Those changes occurred from 3 mutations in the DNA that encoded for the PBP1A-1 protein. The changes were evaluated in relation to the sequence of H. pylori strain 26695 that was susceptible to amoxicillin. The F125L mutation was observed in 6 of the isolates, whereas the I79V mutation was observed in 4 of the isolates (Table 5).
Description of the mutations in DNA in the pbp1a-1 sequences in the H. pylori isolates.
| H. pylori 26695 | Change | ||||||
|---|---|---|---|---|---|---|---|
| Codon-i | AAi | DNA | Codon-f | AAf | Mutation | R | % |
| TTT | F | T373C/T375C | CTC | L | F125L | 6 | 60 |
| ATT | I | A235G | GTT | V | I79V | 4 | 40 |
| Total | 10 | 100 | |||||
AAf: amino acid encoding for codon-f; AAi: original amino acid encoding for codon-I; Codon-f: sequence of the mutated codon in the resistant H. pylori strains; Codon-i: sequence of the original codon in H. pylori strain 26695; R: number of mutations observed in the sample.
Amoxicillin is a commonly used antibiotic in H. pylori eradication therapy. Even though the majority of H. pylori isolates are still susceptible to amoxicillin, resistance to that antibiotic is emerging in clinical isolations, especially in countries where the drug is widely distributed without a prescription.16 At present, the underlying molecular mechanism to resistance to amoxicillin has only been identified in a few H. pylori isolates. In those isolations, resistance was mediated by mutations in the gene that encodes PBP1A.16
The PBPs are a group of proteins that are characterized by their affinity for and binding to penicillin. They are enzymes that participate in the synthesis and maintenance of the peptidoglycan layer of the bacterial cell wall.16 It is important to keep in mind that there are 2 types: PBP1A and PBP2A.7 Resistance is produced through modifications in the PBPs, which leads to a decrease in the affinity for the medication. Those modifications include mutations and/or mosaics in PBP2X and PBP2B, as well as in PBP1A, for the highly resistant isolates.14
There are few Colombian studies directed at evaluating H. pylori susceptibility to amoxicillin and they differ in the susceptibility analysis methodology and the prevalence of resistance. In our study, the prevalence of resistance to amoxicillin in H. pylori isolates from a Túquerres population was 5.4% (8/147), which is very similar to values reported in previous studies. In Bogotá, the proportion of resistance was reported at 7% (6/84), through the disk diffusion technique,10 and at 3.8% (3/79), through the E-test technique,11 and contrasted with that reported in H. pylori isolates from the coffee-growing region (Armenia and Pereira), where resistance to amoxicillin did not occur.15 Those registers illustrate the geographic variation in the prevalence of resistance to amoxicillin in Colombia. The contrasts in said prevalence could be explained by greater exposure to the antimicrobial.17,18
Variants and changes were found in the uncharged polar amino acids, F125L and I79V. The association of those mutations could be explained by their close presence to the enzymatic activity sites of the PBP protein that are located in the C-terminal region (SKN368-371, SNN433-435, and KTG555-557). That is where mutations cause conformational changes in the PBPs, decreasing the binding affinity to amoxicillin and resulting in resistant phenotypes.19 On the other hand, the association could also be explained by the presence of mutations, such as A296V, A494H, A541M, and Q572G, in the pbp2 gene, and A499V and Q536L in the pbp3 gene, which are found in regions not evaluated in the present study.20,21
Despite the fact that the mutations in the pbp1a gene are associated with resistance to amoxicillin in H. pylori, there are other genes that could be responsible for the resistant phenotype, such as mutations in the hopC gene and the deletion in the porin-encoding hopB gene of H. pylori, whose presence enables the bacterium to survive at amoxicillin concentrations of 125mg/l and 250mg/l, respectively.22 In addition, if the changes are simultaneous to mutations in the pbp1a gene, the bacterium can grow in amoxicillin concentrations up to 400mg/l, suggesting an additive effect.23 Beta-lactamases are among the factors associated with resistance, as described in a study on H. pylori strain 3778, in which a product identical to beta-lactamaseTEM-1 (GenBank accession: EU726527) was found. It enabled the growth of that strain in media with amoxicillin concentrations above 256mg/l.24 Efflux pumps are not likely involved in resistance to amoxicillin, given that the antibiotic has very low hydrophobicity, which is an indispensable requisite in drugs that are substrates of those types of structures.25
The present study is the first report on mutations in the pbp1a gene in H. pylori isolates from a Túquerres population. Mutations were found that have not been reported in previous analyses. Despite our study results, we suggest evaluating the effect of the mutations on the designs of directed mutation or modeling, to identify their true contribution to resistance to amoxicillin.
Study strengths and limitationsThe aims of the present study were to characterize the mutations present in the pbp1a gene and to determine their possible association with resistance to amoxicillin in vitro, in a patient population from Túquerres, located in the Colombian Andes; the prevalence of H. pylori (>80%); and the high risk for the development of gastric cancer. The use of microbiologic and molecular techniques to identify H. pylori provided more reliable results, but studies are needed to evaluate isolates from different regions of Colombia and the world, to identify mutations in the strains circulating in the environment and their association, not only with resistance to amoxicillin, but to other drugs used in H. pylori eradication, to reduce treatment failure.
Financial disclosureThis project was financed by the Departamento Administrativo de Ciencia Tecnología e Innovación de la República de Colombia - COLCIENCIAS, Grant No. RC: 1902002; and by the Registro Poblacional de Cáncer de Cali, Universidad del Valle, Cali, Colombia.
Conflict of interestThis project was financed by the Departamento Administrativo de Ciencia Tecnología e Innovación de la República de Colombia - COLCIENCIAS, Grant No. RC: 1902002; and by the Registro Poblacional de Cáncer de Cali, Universidad del Valle, Cali, Colombia.
The authors wish to thank the Microbiology and Molecular Biology Laboratory and the Histopathology Laboratory of the Pathology Department of the Universidad del Valle for the use of their installations for the development of this study, as well as the Hospital San José of Túquerres for the use of their installations for taking the clinical samples from which the H. pylori isolates used in this study were obtained.
In addition, they wish to thank Colciencias and the Universidad del Valle for supporting and financing the present study, and lastly, to thank the research group of the Registro Poblacional de Cáncer de Cali research group and its members.
Please cite this article as: Matta AJ, Zambrano DC, Martínez YC, Fernández FF. Mutaciones puntuales en el dominio transglicosilasa del gen pbp1a en aislados de Helicobacter pylori resistentes a amoxicilina. Revista de Gastroenterología de México. 2023;88:100–106.