Colorectal cancer (CRC) is the third most prevalent cancer worldwide. Many risk factors are involved, and current evidence links the gut microbiota and colorectal carcinogenesis. Fusobacterium nucleatum (F. nucleatum) is proposed as one of the risk factors at the onset and during the progression of CRC, due to immune system and inflammatory modulation.
Materials and methodsNinety samples from three different regions of the colon were collected through colonoscopy in patients with CRC, and qPCR TagMan® was conducted to detect F. nucleatum and cytokines (IL-17, IL-23, and IL-10) in tumor, peritumor, and normal samples. The differences between them were analyzed and correlated.
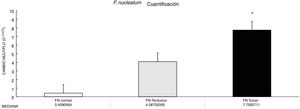
ResultsThe abundance of F. nucleatum determined through the 2-ΔΔCt method in CRC (7.750 [5.790-10.469]) was significantly higher than in the normal control (0.409 [0.251-0.817]) (p < 0.05). There was no significant association between F. nucleatum and the cytokines (p > 0.05).
ConclusionsCRC is a heterogeneous disease that presents and progresses in a complex microenvironment, partially due to gut microbiome imbalance. F. nucleatum was enriched in CRC tissue, but whether that is a cause of the pathology or a consequence, has not yet been clearly defined.
El cáncer colorrectal (CCR) es el tercer cáncer más prevalente en el mundo. Son muchos los factores de riesgo y la evidencia actual apunta a una conexión entre la microbiota intestinal y la carcinogénesis colorrectal. Se propone al Fusobacterium nucleatum (F. nucleatum) como uno de los factores de riesgo en el inicio y la progresión del CCR, por modulación inmune e inflamatoria.
Materiales y métodosSe recolectaron 90 muestras de tres diferentes regiones del colon por medio de colonoscopía en pacientes con CCR y se realizó qPCR TagMan® para detectar F. nucleatum y citocinas (IL-17, IL-23 e IL-10) en el tumor, a nivel peritumoral y en tejido normal. Las diferencias entre las muestras fueron analizadas y correlacionadas.
ResultadosLa abundancia de F. nucleatum determinada por medio del método 2-ΔΔCt en CRC [7.750 (5.790 - 10.469)] fue significativamente mayor que el control normal [0.409 (0.251 - 0.817)] (P < 0.05). No se observó asociación significativa entre F. nucleatum y las citocinas (p > 0.05).
ConclusionesEl CCR es una enfermedad heterogénea que se presenta y progresa en un microambiente complejo, parcialmente debido a desequilibrio en la microbiota intestinal. La F. nucleatum se encuentra de manera abundante en los tejidos del CCR, pero no está aun claramente definido si esto es casual o consecuencia del CCR.
Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive tract, and is currently deemed a major public health concern.1 Overall, colorectal cancer ranks fourth in incidence (9.2% of the total cancer cases), but second in terms of mortality (9.2% of the total cancer deaths).2 In Mexico, there has been an upward trend in mortality rates from the disease over the past three decades. CRC is ranked among the 10 most prevalent causes of morbidity from malignancies in Mexico.3 Colorectal carcinogenesis is a heterogeneous process associated with various sets of somatic molecular alterations that are influenced by diet, environment, and microbial exposures. Inflammation has been identified as an important risk factor.4 The human digestive tract has over 1014 bacteria, eukaryotes, and viruses that form the so-called gut microbiota. Those microorganisms play a significant role in normal human physiologic activities, including digestion, metabolism, epithelial homeostasis, and gut lymphoid tissue development. Dysbiosis in the gut microbiota, such as changes in its population or composition, can cause specific diseases (e.g., cardiometabolic disorders, inflammatory bowel disease, neuropsychiatric diseases, and cancer).5 Metagenomic analyses involving whole-genome sequencing, transcriptome sequencing, and 16S ribosomal RNA gene DNA sequencing have recently demonstrated F. nucleatum enrichment in CRC tissue, compared with control groups. 6–8 Moreover, F. nucleatum had a close relationship with the poor prognosis of CRC patients and most likely promoted chemoresistance.9,10
F. nucleatum is an anaerobic, gram-negative bacillus, present in species-specific reservoirs in the human mouth, gastrointestinal tract, and other parts of the body. It is now well-established that chronic inflammation induced by bacterial infection increases the risk of cancer.11 A recent study has provided insights into the relation between the gut microbiome and inflammatory cytokine production capacity.12F. nucleatum induces local inflammatory cytokines, including IL-6, IL-8, IL-17, TNF-α, and COX-2, in the tumor microenvironment, and those cytokines can promote tumorigenesis in CRC. 6,13–15 The aim of our study was to evaluate the presence of F. nucleatum and its relation to local inflammation in tumor and non-tumor tissue from Mexican CRC patients.
Materials and methodsA total of 30 consecutive patients (18 men and 12 women, age range: 45-84 years) that presented with CRC, histologically confirmed as adenocarcinoma, underwent colonoscopy at the Department of Colorectal Surgery of the Hospital Militar in Mexico, within the time frame of March-September 2019. Patients with colorectal tumors other than adenocarcinoma, those that received chemotherapy or radiotherapy before colonoscopy, and patients that had comorbid malignancies from other organs were not included. Fresh tumor (a macroscopic lesion) tissue samples, peritumor (close to a macroscopic lesion) tissue samples, and control mucosa samples (10 cm beyond the cancer margins) were collected from each subject. The tissue and control mucosa samples were snap frozen in liquid nitrogen for long-term storage at −80 °C, until use. All patients were enrolled in the qPCR study.
DNA extractionDNA extraction from the colorectal tissue samples was performed using the TRIzol reagent (Ambion, Carlsbad, CA, USA) and the manufacturer’s corresponding protocol. Complementary DNA (cDNA) synthesis was carried out using the miScript II RT kit® (Qiagen, Hilden, Germany), with the Veriti Thermal Cycler (Applied Biosystems), following the manufacturer’s instructions.
Quantitative real-time polymerase chain reactionOnce cDNA was obtained, the tests were equalized in each sample through dilutions, to form groups and carry out the amplification and sequencing of the bacterial gene, 16S rRNA.16 The V3 and V6 hypervariable regions of the 16S rRNA gene were amplified by PCR from microbial genomic DNA with the forward (TATGGTAATTGTGTGCCAGCMGCCGCGGTAA) and reverse (GGACTACHVGGGTWTCTAAT) primers. The primers were designed with protruding adapters (forward: AATGATACGGCGACCACCGAG), (reverse: CAAGCAGAAGACGGCATACGAGAT), according to the MiSeq™ System, Illumina, Inc. (San Diego, CA, USA), to recognize the Illumina index sequencing adapters that were added in a subsequent PCR. The PCR products were evaluated by 2% agarose gel electrophoresis and then purified. After purification, the PCR products were quantified through spectrophotometry. The samples were normalized to a final concentration of 2 nM. The 16S rRNA libraries were prepared, using a two-step PCR protocol: First, the extracted DNA concentrations were quantified and then all DNA samples were diluted to the concentration of the most diluted DNA sample, and 2 μl of each DNA sample were used for a PCR reaction at 98 °C for 30 s (98 °C for 30 s, 52 °C for 30 s, 72 °C for 30 s) for 20 cycles, and a 4 °C hold. For each DNA sample, four PCR reactions of 25 μl were performed, which were then pooled and cleaned with AmpureXP microspheres. The second PCR reaction, 4 μl from the previous PCR product, were used with the PE-PCR-III-F and PE-PCR-IV-barcode primers in four reactions of 25 μl, with a 98 °C PCR cycle for 30 s (98 °C for 30 s, 83 °C for 30 s 72 °C for 30 s) for 7 cycles, and a 4 °C hold. Each set of four PCR reactions was pooled and the reactions were cleaned up, using 16S metagenomic sequence purification beads. The library concentrations were quantified. The library was sequenced on a single set of Illumina MiSeq lines, for the base pair readings of 300 pairs.
F. nucleatum and cytokine levels were given as relative quantification and determined by 2-ΔCt, where ΔCt was the difference in the Ct number for the test and reference (Glyceraldehyde-3-phosphate dehydrogenase[GAPDH]) gene assay. The fold increase of F. nucleatum and cytokine quantification in the diseased tissue over the matched normal colorectal tissue was calculated as 2-ΔΔCt. Previously published primers and probes17–19 were synthesized by the Integrated DNA Technologies Company (Coralville, Iowa, USA), with the following sequences:
- 1)
F. nucleatum: forward primer, 5′ CGC AGA AGG TGA AAG TCC TGT AT 3′. reverse primer, 5′ TGG TCC TCA CTG ATT CAC ACA GA 3′.
- 2)
IL-17: forward primer, 5′- CTC ATT GGT GTC ACT GCT ACT G - 3′. reverse primer, 5′ - CCT GGA TTT CGT GGG ATT GTG - 3′.
- 3)
IL-23: forward primer, 5′- ACA CAT GGA TCT AAG AGA AGA GG - 3′. reverse primer, 5′- CTA TCA GGG AGC AGA GAA GG - 3′.
- 4)
IL-10: forward primer, 5′- AAT AAG GTT TCT CAA GGG GCT - 3′. reverse primer, 5′- AGA ACC AAG ACC CAG ACA TCA A - 3′
- 5)
GAPDH: forward primer 5′- GTCTCCTCTGACTTCAACAGCG -3′. reverse primer 5′- ACCACCCTGTTGCTGTAGCCAA – 3′
All statistical analyses were performed using the SPSS 25.0 for Windows (SPSS Inc. Chicago, IL, USA). The relation of F. nucleatum to cytokine levels between tumor tissue, peritumor tissue, and matched normal colorectal tissue was given as fold increase 2−ΔΔCt, where ΔΔCt was the median of the difference between ΔCt Diseased Tissue and ΔCt Normal Tissue. Continuous data were expressed as medians (25ᵀᴴ percentile, 75ᵀᴴ percentile). A test of normality was carried out using the Shapiro-Wilk test. Due to the non-Gaussian nature of the qPCR data distribution, non-parametric testing was deemed appropriate. Consequently, the Wilcoxon rank sum test was used to compare the median F. nucleatum levels and cytokines in the diseased tissue versus the matched normal tissue and to compare the F. nucleatum levels between the colon subsites. The Spearman rank correlation coefficient test was performed to compare F. nucleatum quantification with cytokine expression. For all analyses, a p value < 0.05 (two tails) was considered statistically significant.
Ethical considerationsThe authors declare that informed consent was obtained from all participating subjects, in accordance with the Declaration of Helsinki, and all samples were coded to protect patient anonymity. The study was approved by the Research Ethics Committee of the Hospital Central Militar, affiliated with the Universidad del Ejército and the Fuerza Aérea de México.
ResultsF. nucleatum was more abundant in CRC tumor tissue versus normal tissue.
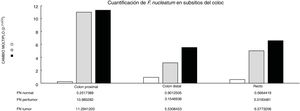
Through qPCR, we were able to confirm that the fold change of F. nucleatum was significantly higher in the diseased tissue (tumor and peritumor samples), regardless of tumor stage and location, compared with the matched normal tissue, in a Mexican cancer cohort (Fig. 1). The details of the clinical data and cancer stage and location are summarized in Table 1. F. nucleatum and cytokine levels were quantified in the 30 CRC samples and in the respective peritumor and normal samples. A significant increase of F. nucleatum for bacterial DNA was found in the tumor samples (7.750 [5.792-10.469]). A paired analysis of F. nucleatum quantification for the tumor group was performed, using normal tissue (0.409 [0.251-0.817]) (Table 2). Different expressions of F. nucleatum levels were shown along the colon, the highest of which was in the proximal colon, with a median fold change of 11.294 and 10.985 in the tumor and peritumor samples, respectively. However, no significant differences between locations were found (p > 0.05) (Fig. 2). Interleukin 23 (IL-23) (0.272 [0.125-0.345]) was significantly reduced in the tumor tissue, compared with the normal samples (1.261 [0.509-1.750]) (Fig. 3). No significant expression of other cytokine levels was found in our study. Table 2 summarizes the tumor and peritumor fold changes for F. nucleatum and the cytokines, compared with the normal samples. The bacterial levels were not significantly correlated with the cytokine levels in the tumor tissue or the peritumor tissue (Table 3).
Clinical data.
| Clinical diagnosis overview | Mexican cohort |
|---|---|
| Total number of tissue samples | 90 |
| Sex, n (male/female) | 18/12 |
| Age, mean ± SD (years) | 65 ± 10 |
| Location, n (proximal colon/ distal colon/ rectum) | 7/11/12 |
| Body mass index, mean ± SD (kg/m²) | 32.5 ± 3.0 |
| Indication for colonoscopy, n (bleeding/abdominal pain/ change in bowel habits/ weight loss) | 12/4/9/5 |
| CEA, mean ± SD (ng/mL) | 25 ± 32 |
| Stage, n (stage I, stage II, stage III, stage IV) | 3/10/11/6 |
CEA: carcinoembryonic antigen; SD: standard deviation.
The fold change (2-ΔΔCt) of F. nucleatum and cytokines in tumor and peritumor tissue samples, compared with normal tissue.
| IL-17 | IL-10 | IL-23 | F. nucleatum | |
|---|---|---|---|---|
| Tumor/normal | ||||
| FC median | 1.741 | 6.536 | 0.272 | 7.750 |
| FC range | 0.632 - 2.875 | 1.383 - 13.336 | 0.125 - 0.345 | 5.792 - 10.469 |
| p value | 0.564 | 0.248 | 0.021* | 0.020* |
| Peritumor/normal | ||||
| FC median | 0.770 | 3.639 | 0.876 | 4.087 |
| FC range | 0.403 - 0.868 | 0.032 - 22.935 | 0.064 - 2.796 | 1.062 - 9.725 |
| p value | 0.564 | 1.000 | 0.564 | 0.081 |
| Tumor/Peritumor | ||||
| FC median | 0.817 | 3.978 | 1.030 | 0.318 |
| FC range | 0.244 - 3.717 | 0.009 - 6.495 | 0.291 - 12.692 | 0.212 - 1.226 |
| p value | 0.309 | 0.564 | 0.773 | 0.248 |
FC: fold change; IL: interleukin.
Wilcoxon sum rank test.
Correlation analysis between the fold change of the cytokines in the normal samples and the fold change of F. nucleatum in the tumor and peritumor samples.
| Spearman correlation coefficient (r) | Tumor | Peritumor |
|---|---|---|
| IL-17 | 0.80 | -0.20 |
| p value | 0.80 | 0.20 |
| IL- 23 | -0.80 | 0.20 |
| p value | 0.20 | 0.80 |
| IL-10 | 0.80 | 0.80 |
| p value | 0.20 | 0.20 |
IL: interleukin.
Spearman correlation test.
*p value < 0.05 was considered statistically significant.
Alterations in the gut microbial composition are thought to play a role in colorectal cancer development.4,5,20 The results of the present study confirm, for the first time in Mexican patients, previous reports from North America, Europe, and Asia6,7,10,21 that F. nucleatum is over-represented in tumor tissue, compared with normal tissue in CRC. Even though there were no significant differences between F. nucleatum levels and tumor location, a high level of the bacterium was observed in the proximal colon. There are well-known differences in clinical, pathologic, and epidemiologic features between proximal and distal colon cancers.22,23 The same differences are seen between the microbiota in the proximal and distal colon. Flynn et al. found that differences in the microbiota of the proximal and distal colon in healthy individuals could be due to the differences in oxygen distribution along the colonic mucosa. The proximal colon primarily hosts aerobic bacteria in its mucosa, and adversely, the distal colon mainly harbors anaerobic species.24 Today, there is growing evidence that various risk factors related to CRC development (smoking, inflammatory bowel disease, high alcohol consumption, excessive consumption of red meat, diabetes mellitus, obesity, and genetic predisposition) greatly affect the synthesis of the gut microbiota.25 Consequently, some bacterial populations are minimized, whereas other bacteria that are rare colonizers of the colon, such as Fusobacterium and Lactococcus, display adaptive behavior to the new environment, thus confirming the hypothesis of a “bacterial driver-passenger” model that plays a key role in CRC pathogenesis.26 Moreover, some studies on bacterial biofilms of the colonic mucosa have provided novel insight into the different pathogenetic mechanisms underlying CRC tumorigenesis between the colonic regions.27 Cancers located in the right colonic regions (cecum, ascending colon, and hepatic flexure) generally demonstrate the substantial presence of biofilms.28 Dejea et al. showed that biofilms were associated with human colon cancer and linked to cancer location, with biofilm positivity in virtually all adenomas and cancers located in the right colonic regions, whereas cancers located in the left colonic regions (distal colon to hepatic flexure) rarely possessed biofilms.28,29 Those bacterial biofilms were related to epithelial changes that characterize the progression of tumorigenesis, including diminished E-cadherin and increased levels of the angiogenic and proinflammatory cytokines.30 Using the “bacterial driver-passenger” model, F. nucleatum has been shown to cause an inflammatory microenvironment that is more favorable for CRC development, in contrast to other bacteria that colonize at the tumor site.6 A persuasive interpretation of the aforementioned data is an important issue. First of all, is F. nucleatum a cause of CRC or a consequence of the disease? So far, increasing evidence is in favor of the “cause” hypothesis, as emerging data have demonstrated that F. nucleatum initially induced precancerous lesions (e.g., hyperplastic polyps and adenomas) that eventually progressed to CRC.9,31 In addition, several pathogenetic studies have supported the carcinogenic role of F. nucleatum, which promotes an oncogenic and inflammatory response via FadA, the major virulence factor of F. nucleatum, binding to E-cadherin and activating the B-catenin pathway.32 Moreover, the presence of F. nucleatum in the gut affects tumor-related cytokines involved in CRC tumor progression.15,33 In the tumor samples, we found that IL-23 was significantly decreased, compared with the normal tissues, whereas there were increases in IL-17 and IL-10, but the difference with normal tissue was not significant (Fig. 1). Because of our study’s cross-sectional nature, the increase in IL-17 lends support to the majority of studies that consider said interleukin an important promoter in tumor initiation and CRC progression, through the IL-23/IL-17 pathway, which has a critical role in CRC pathogenesis.34–36 In our study, there was a decrease in IL-23, leading us to suppose that its reduction during tumor development was due to the fact that it functions as a starter of the IL-23/IL-17 pathway, resulting in an increase in IL-17. Grivennikov et al. demonstrated that interleukin 23 signaling promoted tumor growth and progression, and the development of a tumoral IL-17 response. IL-23 is mainly produced by tumor-associated myeloid cells that are likely to be activated by microbial products.37 Recently, Kostic et al. reported that F. nucleatum selectively expanded myeloid-derived immune cells in colorectal cancer. During tumor progression, reactive myeloid cells could possibly mediate immunosuppression by interleukin 10 (IL-10) production.6 IL-10 is known to be a potent anti-inflammatory cytokine.38 Due to its immunosuppressive effect on dendritic cells and macrophages, IL-10 can dampen antigen presentation, as well as cell maturation and differentiation, allowing tumor cells to evade the immune surveillance mechanism.39 Some studies have shown that miR-21 produced by F. nucleatum increased the levels of IL-10 and prostaglandin E2 (PGE2) in colorectal cancer cells,40 and McCoy et al. found a significant positive correlation between F. nucleatum abundance and IL-10 expression in rectal mucosa biopsies from adenoma patients, compared with normal controls.31 Our results suggest an expansion of F. nucleatum colonization in CRC tissues, a phenomenon that could lead to increased proinflammatory mediators (cytokines), such as IL-10. A study by Proença et al. examined the influence of F. nucleatum in colorectal adenoma and CRC on inflammatory mediator expression through miRNA activation that contributed to colorectal carcinogenesis.41 If validated, F. nucleatum DNA in colorectal carcinoma tissue could be a prognostic biomarker. In addition, our study might provide insights for future studies to develop strategies for colorectal cancer prevention and treatment, through targeting the microbiota.
The present study has several notable strengths and limitations. Its strengths include the use of DNA extracted from frozen fresh tissue, with no tissue fixation, resulting in > 99% successful qPCR assays (based on consistent amplification of the human GAPDH reference gene in all tumor samples, peritumor samples, and matched normal samples), and the fact that we assessed the prognostic utility of F. nucleatum DNA measurement in CRC patients, reflecting the main features in Mexican patients. An important limitation was the low number of patients included in the study (n = 30). Testing should be carried out on a larger sample, and more intestinal bacterial species, in addition to F. nucleatum and cytokines, should be analyzed, as well.
ConclusionThe present study supports the evidence that the presence of F. nucleatum, as a symbiotic bacterium in the human intestinal tract, is related to the development of CRC. F. nucleatum is apparently stimulated in the proximal colon to promote CRC through different virulence mechanisms, such as adhesion to the intestinal epithelium and induction of inflammatory and immune responses in the host. Among those responses are cytokine synthesis and release, including activation of the IL-23/IL-17 signaling pathway, which leads to CRC progression. Therefore, F. nucleatum plays an important role in colorectal carcinogenesis and could be targeted for colorectal cancer prevention and treatment in the future.
Financial disclosureThe authors declare that no funding was received in relation to this research project.
Author contributionsH. Cuellar-Gomez designed and performed the research and drafted the manuscript, M.E. Ocharan-Hernandez designed the research and contributed to the analysis, and C. C. Calzada-Mendoza and D.A. Comoto-Santacruz supervised the research and contributed to the analysis.
Conflict of interestThe authors declare that they have no conflicts of interest.
H Cuellar-Gomez gratefully acknowledges the scholarship from the CONACyT to pursue his postgraduate studies.
Please cite this article as: Cuellar-Gómez H, Ocharán-Hernández ME, Calzada-Mendoza CC, Comoto-Santacruz DA. Asociación entre infección por Fusobacterium nucleatum y cáncer colorrectal: un estudio mexicano. Rev Gastroenterol México. 2022;87:277–284.